Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Similares em
SciELO
Similares em
SciELO
Compartilhar
Portugaliae Electrochimica Acta
versão impressa ISSN 0872-1904
Port. Electrochim. Acta vol.36 no.1 Coimbra jan. 2018
https://doi.org/10.4152/pea.201801001
Electrochemical Aptasensor Array for Multiple Detection of Human Osteopontin
Sofia G. Meirinhoa,b,* , Luis G. Diasc,d , Antonio M. Peresb and Ligia R. Rodriguesa
a CEB - Centre of Biological Engineering, University of Minho, Gualtar Campus, 4710-057 Braga, Portugal
b LSRE-LCM - Laboratory of Separation and Reaction Engineering - Laboratory of Catalysis and Materials, ESA, Polithecnic Institute of Braganca, Santa Apolónia Campus, 5300-253 Braganca, Portugal
c ESA - Polithecnic Institute of Braganca, Santa Apolonia Campus, 5300-253 Braganca, Portugal
d CQ-VR - Centre of Chemistry Vila Real, University of Tras-os-Montes, 1013 P.O. box, 5001-801 Vila Real, Portugal
Abstract
Electrochemical aptasensor arrays have been recently used to detect several proteins reported as disease biomarkers, mainly due to their known advantages, that include high affinity, sensitivity, specificity and low costs, among others. This study describes the development of a label-free electrochemical multi-aptasensor array, for the simultaneous detection of human osteopontin (OPN), using two specific aptamers. To enable multiplexed protein assay, RNA and DNA aptamers were immobilized in the dual-screen-printed gold working electrodes via streptavidin-biotin interaction, and using the [Fe(CN)6]3-/4- as the redox probe for cyclic voltammetry measurements. The multi-aptasensor array herein developed exhibited a good response and selectivity to detect human OPN in the presence of other interfering proteins. Considering these preliminary results, the DNA/RNA dual aptasensor array could potentially be used as an analytical tool for the specific detection of human OPN, and for cancer diagnosis overall.
Keywords: ptasensor array, electrochemical detection, osteopontin, aptamer, dual- screen-printed gold electrode.
Introduction
The simultaneous detection of multiple proteins or the multiple detection of a specific protein in a single assay, by using biosensor devices with several working electrodes (WE), open new promising possibilities regarding the design of new and personalized approaches that may be used for the early detection and therapy of many diseases, e.g. cancer [1-3]. Most cancer diseases are associated with the presence of several biomarkers, such as proteins that could be used as potential diagnostic or prognostic markers, indicative of the disease stages [4, 5]. Osteopontin (OPN) is a phosphorylated glycoprotein that has been reported as a potential breast cancer biomarker. Indeed, its over expression may be indicative of tumour formation, cancer progression, metastasis and poor prognosis [6-9]. To improve the survival of patients with cancer diseases, their early diagnosis is crucial. Therefore, it is necessary to develop new, simple and cost-effective methods for the detection and quantification of these protein biomarkers. The aptasensor arrays could be an effective alternative to conventional analytical methods, considering their known advantages, such as the possibility of simultaneous multiple detection of proteins in a single assay, the reduction of the sample and reagents' volumes, and of the analysis' costs, and the possibility to perform several replicates per assay [3, 10-12]. The development of aptasensors arrays requires the design of screen-printed electrodes (SPE) with several working electrodes, and the selection of aptamers in similar experimental conditions (e.g. pH, temperature, length of the sequence, ionic changes, among others), aiming to minimize the risk of affecting their high affinity and specificity towards the target molecule [10,11].
The simultaneous detection of multiple protein biomarkers using multi-electrodes arrays based on immunosensors has been reported in the literature [13,14]. In the last years, the use of electrode arrays to develop novel electrochemical aptasensor devices has gained popularity. Song et al. [4] described an electrochemical aptasensor array for the multiplexed detection of platelet-derived growth factor-BB (PDGF-BB) and thrombin using a SPE array with four subarrays, four independent carbon working electrodes (WE) and one counter electrode (CE). The proposed aptasensor array allowed multi-detection with good selectivity, showing both PDGF-BB and thrombin (THR) aptamer-modified electrodes only stripping responses against their respective proteins. Moreover, no cross-reaction between the two aptamers towards non-cognate proteins was reported. Liu et al. [15] described an electrochemical aptasensor array for the detection of interferon gamma (IFN-γ) and tumour necrosis factor alpha (TNF-α), using micropatterned aptamer-modified gold electrodes. The developed aptasensor array showed similar sensitivity to that of the single aptasensors for both proteins, and it was found to be applicable for the detection of those proteins in activated T cells and monocytes cell lines. Recently, the same research group [16] reported an electrochemical aptasensor for the specific detection of multiple cytokines (IFN-γ and TNF-α), using an electrode-array with a set of four miniaturized WE. The aptasensor showed high specificity and sensitivity, similarly to that of the single analyte aptasensor, for each biomarker. This work describes the development of an electrochemical aptasensor array using two specific aptamers for the simultaneous detection of human OPN. Although aptasensors are usually used for the multiplex analysis of different proteins, they can also be used to increase the sensitivity of detection of a single protein. Thus, in the current work, two different aptamers were used to detect and quantify OPN. For that purpose, a dual-screen-printed gold electrode (dual- SPGE) was used.
Materials and methods
Reagents and apparatus
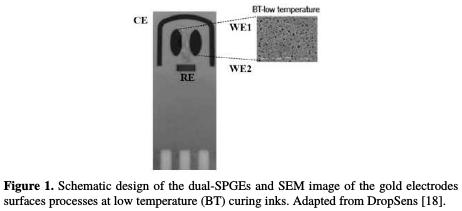
Diethylpyrocarbonate (DEPC), 3,3-dithiodipropionic acid (DPA), N-(3- dimethylaminopropyl)-N-ethylcarbodiimide hydrochloride (EDC), N-hydroxysuccinimide (NHS), ethanolamine (ETA) and sulfuric acid (purity of 99.999 %) were obtained from Sigma-Aldrich. Potassium hexacyanoferrate (III) (K3Fe(CN)6) and potassium hexacyanoferrate (II) (K4Fe(CN)6) were obtained from Acros Organics, and potassium dihydrogen phosphate (KH2PO4) from Merck. Sodium chloride (NaCl), potassium chloride (KCl) and sodium hydrogen phosphate (Na2HPO4) were acquired from Panreac. All chemicals were of analytical grade and used as received. All proteins were acquired under a lyophilized form, and prepared according to the manufacturer specifications. The stock solutions of each protein were stored at -20 °C. The protein working solutions were obtained by dilution of the stock solutions with phosphate buffer saline (PBS) (pH 7.4), and were stored at 4 °C until use. Recombinant human OPN (rhOPN, 65 kDa) and recombinant bovine OPN (rbOPN, 60 kDa) were purchased from R&D Systems. Thrombin from human plasma (THR, 37.4 kDa), bovine serum albumin (BSA, 66 kDa), lysozyme from chicken egg whites (LYS, 14.3 kDa) and streptavidin were obtained from Sigma-Aldrich. Figure 1. Schematic design of the dual-SPGEs and SEM image of the gold electrodes surfaces processes at low temperature (BT) curing inks. Adapted from DropSens [18]. The RNA aptamer (R3-OPN, described by Mi et al., [17]) and DNA aptamer (C10K2, selected and developed by our research group (work under revision)) were synthesized by Integrated DNA Technologies (Belgium). Their sequences were as follow: RNA aptamer (5'-Biotin- CGG CCA CAG AAU GAA AAA CCU CAU CGA UGU UGC AUA GUU G-3') and DNA aptamer (5'-Biotin- TGT GTG CGG CAC TCC AGT CTG TTA CGC CGC-3'). Stock solutions of DNA and RNA aptamers (100 μM) were prepared with ultra-pure water containing 1 % (v/v) of DEPC, to avoid the RNase interference. The working DNA and RNA aptamer solutions were prepared by diluting the stock solutions using fresh PBS. The binding recognition between both aptamers and OPN was studied by cyclic voltammetry (CV), using a Potentiostat-Galvanostat device (PG580, Uniscan Instruments). The pH was measured using a pH meter (iHANNA instruments pH 211). The dual-SPGEs were purchased from DropSens (Oviedo, Spain). These dual-SPGEs include a four-electrode system configuration printed on the same strip of ceramic substrate (3.4 Ã 1.0 Ã 0.05 cm), and were subjected to low temperature (BT) ink curing. The dual-SPGEs were composed of two parallel ellipses of gold-BT WE (WE1 and WE2 with a surface of 6.3 mm2 each) located at the ceramic strip, an Ag/AgCl reference electrode (RE) and a gold-BT counter electrode (CE) (19.8 mm2 and 1 mm wide), as shown in Fig. 1.
Immobilization of aptamers onto the dual-SPGEs
The aptamers' immobilization onto the dual-SPGE was achieved through the streptavidin-biotin interaction, as reported by Meirinho et al. [19]. Fig. 2 shows the aptamers' immobilization onto the two gold WE.
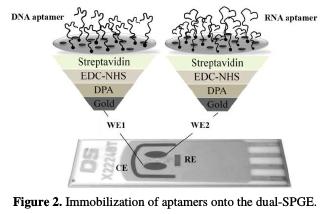
First, the dual-SPGE was cleaned using three acid solutions (0.5 M H2SO4, 0.01 M KCl/0.1 M H2SO4 and 0.05 M H2SO4) under an electric potential, in the range of -0.3 V to +1.5 V, and at a scan rate of 100 mV/s. Afterwards, the gold WEs surfaces were modified using 200 mM of DPA (30 min at room temperature), 100 mM of EDC and 1 mM of NHS (1:1 v/v, 60 min at room temperature), 1 mg/mL of streptavidin (overnight at 4 °C), and 100 mM of ETA (20 min at room temperature). Finally, the biotinylated aptamers were immobilized onto the gold WE. A solution of each biotinylated aptamer (DNA and RNA, 4 nM) prepared in PBS buffer (pH 7.4) was subjected to a temperature treatment (95 °C during 5 min, 4 °C for 5 min and 10 min at room temperature) to obtain adequate suitable structure flexibility of the aptamers, allowing their interaction with the streptavidin on the gold electrode surface. After this pre-treatment step, the DNA aptamer and RNA aptamer were incubated into the WE1 and WE2, respectively, during 40 min at room temperature, being bound to the surface through the streptavidin-biotin interaction.
Evaluation of the aptasensor array specificity towards OPN
The aptasensors array performances, concerning the rhOPN detection, as well as their specificities, were evaluated by CV assays. For the detection of rhOPN, the dual-SPGE modified with DNA aptamer (WE1) and RNA aptamer (WE2) were incubated with 10 μL of an rhOPN solution (200 nM) during 60 min at room temperature. To remove the non-specifically bound protein, the electrodes surfaces were washed with PBS buffer. The specificity of the arrays was evaluated using individual non-specific proteins (BSA, THR, LYS and rbOPN) instead of rhOPN, in the detection experiments with dual-SPGE. This approach has been previously reported [20, 21]. The aptasensors array response towards non-specific binding proteins was assessed using different dual-SPGE modified with DNA aptamer and RNA aptamer into the WE1 and WE2, respectively, which were incubated under the same experimental conditions tested for rhOPN (i.e. 10 μL of each interfering protein solution at 200 nM).
Electrochemical measurements
In all CV measurements, 100 μL of the [Fe(CN)6]3-/4- solution (redox probe) were applied on the electrochemical reaction area of the dual-SPGE electrode. The CV measurements were performed under a potential range of -0.5 V to + 0.8 V, and at a scan rate of 50 mV/s. The decrease in the peak current intensities was expressed as relative current change (ΔI %). The ΔI % was calculated considering the current values of the cyclic voltammogram oxidation peak recorded after aptamer immobilization and protein incubation, by using the equation (1):
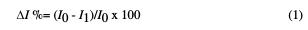
where ΔI is the relative current change (%); I0 and I1 represent the current before and after the sample incubation, respectively.
Results and discussion
Characterization of the dual-SPGE
The purpose of this work was to develop an electrochemical aptasensor array to detect rhOPN simultaneously using two different aptamers (RNA and DNA aptamers). The experimental conditions were previously established by Meirinho et al. [19]. Hence, the aptamer concentration to immobilize it onto the gold WE (WE1 and WE2) and the incubation time for the aptamer-rhOPN interaction were fixed at 4 nM and 60 min, respectively. The steps followed during the aptasensor array construction and electrode surface functionalization were monitored by CV. This electrochemical technique is simple and sensitive, allowing to acquire information concerning the reversibility of the oxidation and reduction mechanisms that occur at the electrode surface [22]. As an example, Fig. 3 illustrates the electrochemical characterization of the aptasensor array, namely: the cyclic voltamograms recorded for the bare Au electrode; the cleaning procedure with acid solution; t and the aptamer immobilization and aptamer- rhOPN binding step, for both aptamers.
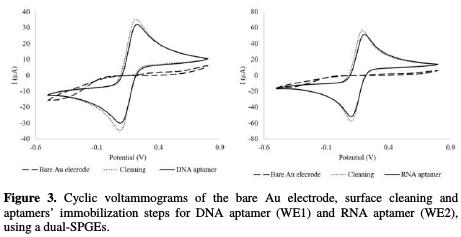
The current responses obtained for the bare Au electrode in both gold WE (WE1 and WE2) were almost null. The almost absence of the current response, and the observation of a quasi-reversible redox peak in the presence of [Fe(CN)6]3-/4-, used as the redox probe, could be attributed to the fact that these electrodes were exposed to a low temperature (BT) curing ink process during their production. After the cleaning step with different H2SO4 solutions, characteristic quasi-reversible voltammograms were obtained for both gold WE, demonstrating that this step was essential for the activation of the gold WE' surfaces (WE1 and WE2), allowing to obtain a uniform and functionalized surface.
The step corresponding to the RNA and DNA aptamers immobilization onto the gold working surfaces showed a decrease of the peak current intensities compared to the cleaning step. This decrease may probably be attributed to the negative charges of the aptamers backbone phosphate group and the [Fe(CN)6]3-/4- redox probe, resulting in electrostatic repulsive interactions that could block the electron transfer [23]. Finally, these results suggest that both aptamers were successfully assembled onto the gold working surface.
Evaluation of the aptasensor array performance
The aptasensors array developed for the simultaneous detection of rhOPN was electrochemically evaluated. After the rhOPN incubation (during 60 min onto the dual-SPGE modified) with the DNA aptamer (WE1) and the RNA aptamer (WE2), significant changes of the voltammograms were observed in comparison with the voltammograms recorded after the aptamers' immobilization, which could be due to the change of the aptamer conformation upon protein binding (Fig. 4).
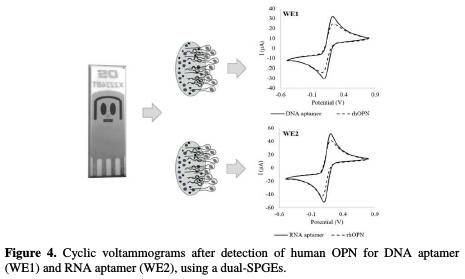
The specific recognition of RNA and DNA aptamers led to the formation of the aptamer-rhOPN complexes, and similar decreases in the current were observed for both gold WE: ΔI (%) of 20% ± 3% for the DNA aptamer-rhOPN and ΔI (%) of 19% ± 2% for the RNA aptamer-rhOPN. These values are within the same order of magnitude as those obtained in the single analyte aptasensor for RNA aptamer [18] and for DNA aptamer [21]. The aptasensors array exhibited a good response for rhOPN, with a negative readout signal upon protein binding to aptamer (i.e. a signal-off sensing mechanism). The observed signal decrease could be even enhanced if several parameters were optimized during the aptasensors' array design. Indeed, it is known that several factors could affect the sensitivity and selectivity of the aptasensors, namely the surface of the electrode (e.g. composition and area), bioreceptors characteristics (e.g. sequence length, experimental conditions isolation/selection) and the experimental conditions used to develop the biosensor (e.g. used buffer, incubation temperature and time used for the aptamers immobilization, as well as each aptamer concentration to immobilize it onto the electrode surface). However, in this study, the used experimental conditions were the ones previously optimized for a gold WE with a much lower surface area (0.5 mm2) [19], which could have conditioned the overall performance herein reported.
The specificity of the electrochemical response of the dual-aptasensors array (DNA and RNA immobilized aptamers) was evaluated through the response to the presence of non-specific proteins (Fig. 5).
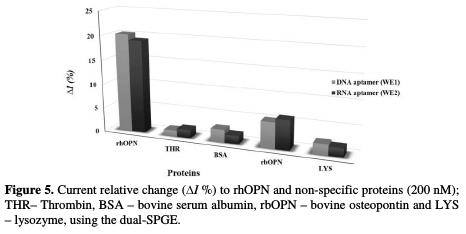
The relative current response (ΔI %) for the tested non-specific proteins was found to be negligible compared to the binding response obtained for rhOPN, with the exception of rbOPN protein. However, since this protein is from animal origin, it is not expected to be present in human biological samples, so this interference does not represent a real concern. Furthermore, the results clearly demonstrated that the dual-aptasensors array may be an interesting complementary tool to detect rhOPN in biological samples, such as blood, plasma or serum.
Conclusions
This study reports the evaluation of a label-free, signal-off aptasensors array for the simultaneous detection of human OPN, which was designed and developed using a dual-SPGE. The results herein reported showed that the dual aptasensors device possesses a good response and selectivity for human OPN. Despite these preliminary encouraging results, the optimization of the aptasensors array for the simultaneous detection of human OPN and/or other protein biomarkers, using different aptamers immobilized onto the electrode array, is required. Finally, the aptasensor performance with synthetic and real biological samples should be evaluated.
References
1. Rusling J F, Kumar C V, Gutkind J S, et al. Analyst. 2010;135:2496. [ Links ]
2. Xie S, Walton SP. Biosens Bioelectron. 2010;25:2663. [ Links ]
3. Xiang Y, Qian X, Jiang B, et al. Chem Comm. 2011;47:4733. [ Links ]
4. Song W, Li H, Liang H, et al. Anal Chem. 2014;86:2775. [ Links ]
5. Misek DE, Kim EH. Int J Proteomics. Article ID 343582. 2011;9:1.
6. Bramwell VHC, Tuck AB, Chapman J-AW, et al. Breast Cancer Res. 2014;16:1. [ Links ]
7. Nassar HR, Namour AE, Shafik HE, et al. Forum Clin Oncol. 2015;6:27. [ Links ]
8. Shevde LA, Samant RS. Matrix Biology. 2014;37:131. [ Links ]
9. Xu Y-Y, Zhang Y-Y, Lu W-F, et al. Mol Clin Oncol. 2015;3:357. [ Links ]
10. Saberian-Borujeni M, Johari-Ahar M, Hamzeiy H, et al. Bioimpacts. 2014;4:205. [ Links ]
11. Touhami A. Nanomedicine. 2015;374. [ Links ]
12. Meirinho SG, Dias LG, Peres AM, et al. Biotechnol Adv. 2016;34:941. [ Links ]
13. Yang Z-H, Zhuo Y, Chai Y-Q, et al. Sci Rep. 2014;4:4747. [ Links ]
14. Zhao C, Wu J, Ju H, et al. Anal Chim Acta. 2014;847:37. [ Links ]
15. Liu Y, Kwa T, Revzin A. Biomaterials. 2012;33:7347. [ Links ]
16. Liu Y, Matharu Z, Rahimian A, et al. Biosens Bioelectron. 2015;64:43. [ Links ]
17. Mi Z, Guo H, Russell MB, et al. Mol Ther. 2009;17:153. [ Links ]
18. DropSens. General Catalogue. 2016;43. [ Links ]
19. Meirinho SG, Dias LG, Peres AM, et al. Biosens Bioelectron. 2015;71:332. [ Links ]
20. Bai HY, del Campo FJ, Tsai YC. Biosens Bioelectron. 2013;42:17. [ Links ]
21. Su Z, Xu H, Xu X. et al. Talanta. 2017;164:244. [ Links ]
22. Farghaly OA, Hameed RSA. Int J Electrochem Sci. 2014;9:3287. [ Links ]
23. Bang GS, Cho S, Kim B. Biosens Bioelectron. 2005;21:863-870. [ Links ]
Acknowledgements
The authors acknowledge the financial support from the Strategic funding of UID/BIO/04469/2013 and from project ref. RECI/BBB-EBI/ 0179/2012 (project number FCOMP-01-0124-FEDER-027462). This work was also financially supported by Project POCI-01-0145-FEDER-006984 - Associate Laboratory LSRE-LCM and by Project UID/QUI/00616/2013 - CQ-VR, both funded by FEDER - Fundo Europeu de Desenvolvimento Regional through COMPETE2020 - Programa Operacional Competitividade e Internacionalizacao (POCI) - and by national funds through FCT - Fundacao para a Ciencia e a Tecnologia, Portugal. S. Meirinho also acknowledges the doctoral grant (ref SFRH/BD/65021/2009) funded by FCT.
*Corresponding author. E-mail address: sgmerinho@ipb.ptm
Received November 10, 2016; accepted January 26, 2017