Introduction
The skin microbiome refers to a diverse community of microorganisms, including bacteria, fungi, and viruses, that reside on the skin’s surface. Most of these microbes are recognized as commensal, playing a vital role in maintaining the overall health and functioning of the skin, although sporadically they may cause skin infections1.
Bacteria are the most abundant type of microorganisms found on the skin, while fungi are the least abundant. This predominance persists even in cutaneous regions, such as the feet, characterized by a high level of fungal diversity. According to the topography and skin conditions, a unique microenvironment can be created into the skin, which can be categorized into three distinct regions: the sebaceous, moist, and dry areas1.
The gut microbiota profile in type 2 diabetes mellitus is similar to that observed in other inflammatory pathologies with subclinical inflammation2. It is expected that in T2D, the skin microbiome has different characteristics from normoglycemic individuals. This holds particular relevance due to the complications of T2D, especially in the foot, where an impaired microbiome may lead to higher prevalence of infections3.
In this review, we intend to clarify if alterations occur in the skin microbiome among individuals with diabetes mellitus (DM) and how these alterations may contribute to specific skin conditions and complications associated with the disease.
Methods
A systematic review, following Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines, was performed using PubMed® and Web of ScienceTM databases on December 1st, 2023, using the Mesh Terms “skin microbiome” AND “ DM” limited to papers written in English and published in the past 10 years. The papers obtained from these searches were combined and duplicates were removed.
All the abstracts from each field were reviewed by two different authors.
The papers retrieved, including reviews and research articles, were considered, while case reports were excluded from the study.
Results
As represented in figure 1, we obtained 50 papers to analyze (22 from PubMed and 28 from ScienceDirect). One duplicate was eliminated, and 20 papers were excluded due to the absence of “skin microbiome” or “diabetes” in the abstract. In addition, one paper was excluded because it was written in German.
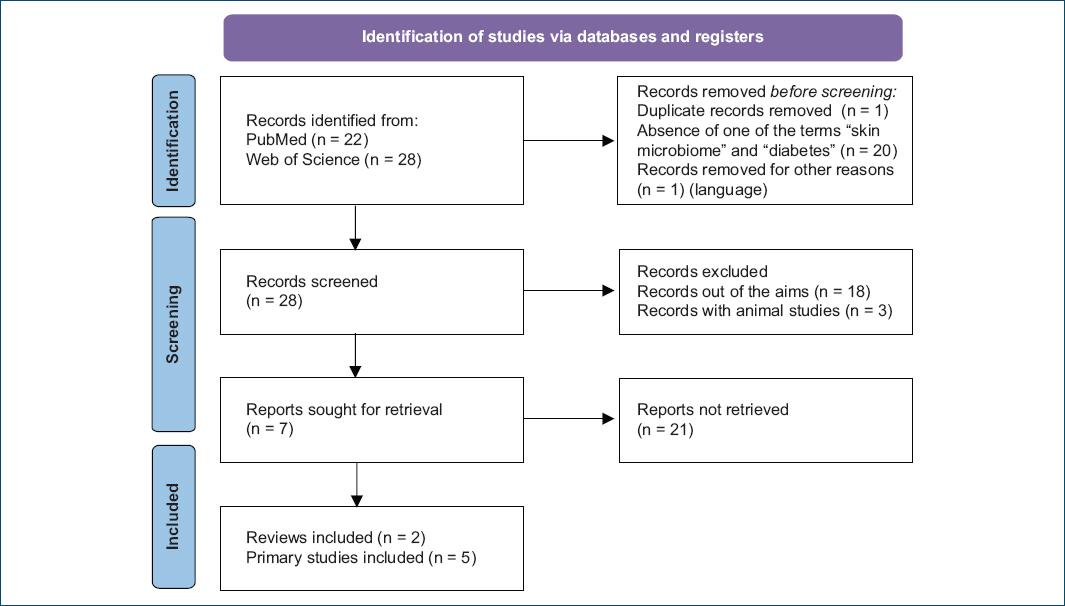
Figure 1 Flow diagram of the literature review using MESH terms “skin microbiome” and “diabetes mellitus”.
After excluding these 22 papers, the authors reviewed the remaining 28 papers. Of these, three did not align with the objective of this review, and three described animal studies.
The remaining papers consisted of two reviews4,5 and five primary studies or research articles6-10, all of which were thoroughly analyzed.
The review led to the conclusion that patients with DM have a higher proportion of Staphylococcus aureus in the skin microbiome than healthy controls4,5. Poor glycemic control was associated with a greater colonization of Staphylococcus spp. and Streptococcus spp5.
One review singled out the top ten bacterial, eukaryotic, and viral species commonly found in various anatomical regions, including dry, moist, sebaceous, and foot skin.
Dry skin in non-diabetic individuals tends to be colonized more frequently by Cutibacterium acnes, Corynebacterium tuberculostearicum, and Streptococcus mitis. Among eukaryotic species, Malassezia restricta, Malassezia globosa, and Aspergillus tubingensis are the most commonly encountered. Moist skin harbors a higher proportion of C. tuberculostearicum, Staphylococcus hominis, C. acnes, as well as fungi like M. globosa, M. restricta, and Tilletia walkeri.
Sebaceous skin is enriched with C. acnes, Staphylococcus epidermidis, and C. tuberculostearicum, along with fungi such as M. restricta, M. globosa, and Malassezia sympodialis.
Notably, S. aureus does not appear among the ten most prevalent bacterial species in any anatomical region representative of dry, moist, or sebaceous skin.
Several primary studies included the analysis of foot samples collected from various sites, including the plantar surface of the foot (both from wound and intact skin, either dry or moist)6, intact skin from the plantar arch of the feet (dry skin)8, the fourth interdigital area of the feet (moist skin)9, chronic skin lesions of the lower limbs (moist and dry skin)7, and both the plantar forefoot (dry skin) and the interdigital space (moist skin)10. However, many of these studies lacked comprehensive descriptions of the features and findings across these different foot areas, hindering proper comparisons. In addition, these studies exclusively focused on patients with type 2 diabetes among individuals with diabetes6-10.
Concerning diabetic foot ulcers (DFUs), the superficial ones, namely, those of short duration, were associated with a predominance of Staphylococcus spp., particularly S. aureus. In contrast, deeper ulcers and those of longer duration exhibited greater microbial diversity, with a higher relative abundance of anaerobic bacteria and Gram-negative Proteobacteria spp. However, the specific period of evolution of these ulcers was not clarified5. Cladosporium herbarum and Candida albicans were identified as the most abundant fungal species found on DFUs. The presence of increased fungal diversity, along with the formation of polymicrobial biofilms consisting of fungi and bacteria, was associated with poor clinical outcomes in chronic wounds5.
Primary cross sectional studies included 8-41 participants in each group (individuals with DM and controls), and studies performed cultural examination, 16S ribosomal RNA sequencing or PCR of the fungal internal transcribed spacer (ITS2) region. These studies revealed that the skin microbiome in individis uals with diabetes, both for bacteria and fungi, was significantly less diverse than in control subjects6-8. Chronic wounds tended to be dominated by the most abundant skin Staphylococcus6,9. A significant association between T2D status and heavy colonization by S. epidermidis (OR-5.40, p = 0.02) was found9.
The bacteriological colony test revealed a higher proportion of both positive bacteriological colony tests and respective polymicrobial result among the individuals with T2D10. This phenomenon confirms an alteration in the skin microbiome of diabetic subjects, indicating a modification in the “opportunistic role” of some species of the skin bacterial flora10.
The 16S rRNA gene sequencing demonstrated dynamic changes in the skin microbiome of the foot during the progression of DM. In patients with DM, the dominant skin microbial phyla were Firmicutes, Proteobacteria, Actinobacteria, and Bacteroidetes7,8.
A single study conducted in China8 investigated the variations in the microbiome among individuals with type 2 DM, without associated complications, across different durations of the disease.
In summary, individuals classified into short-term DM (< 2 years), middle-term DM (5-8 years), and long-term DM (more than 10 years) categories exhibited notable alterations in the microbial community structures of their foot skin compared to the control group (without DM). Moreover, these changes were found to be positively correlated with the duration of the illness8.
At the onset of the disease (short-term DM group), there was a slight reduction in the abundance of Proteobacteria, which steadily increased as the disease progressed (long-term DM group). Conversely, the abundance evolution of actinobacteria displayed an opposite pattern. In addition, the diversity of low-abundance microbes increased with disease progression. All three DM groups demonstrated higher microbial diversity compared to the control group, with the long-term diabetes group exhibiting the highest diversity among the short- and middle-term groups8. Regarding fungi, Trichophyton rubrum was more abundant in DM samples in, exhibiting a lower Shannon diversity index for fungi8.
Discussion
Reviews have underscored the direct involvement of microorganisms in regulating the skin’s immune response.
It is speculated that S. aureus colonization in individuals with DM predisposes them to minor-to-moderate foot infections and even life-threatening bloodstream S. aureus infections, throughout the skin inflammation and an immune response. The role of S. aureus juxtaposes to that in a model of atopic dermatitis, in which S. aureus cutaneous colonization could elicit skin inflammation and induce an immune response4. The proposed mechanisms involves the activation of an inflammatory cascade, intensified by the exposure to S. aureus on the skin surface, which promotes IL-36a production by keratinocytes (partly through the activity of PSMa), which triggers IL-36R/MyD88 signaling on T cells to produce IL-17A/F. In addition, it is suggested that colonization with S. aureus could impair the suppressive activity of Treg cells and staphylococci have the ability to produce lipoprotein acids that can inhibit skin inflammation through a TLR-dependent pathway. The inhibition of complement component C5a receptors reduces the diversity of the skin microbiota, while symbiotic flora can regulate the expression of certain complement genes in the skin, thereby modulating immunity4.
A shared observation across these primary studies is the loss of microbiologic diversity in DM and the increased risk of developing skin infections associated with microbiome dysbiosis.
It was suggested that microbiome dybiosis in T2D could stem from the same activated innate immune response thought to be central to the development of T2D. The extent to which alterations in the microbiome at one organ site influence distal organs or different organ sites remains unclear. In addition, it is uncertain whether these systemic effects are specific to particular tissues or organs, along with the underlying mechanisms involved9.
For the reasons mentioned so far, targeted microbiome modulation reveals such a promising candidate that was recently discovered to exert anti-inflammatory and beneficial metabolic functions, that can mitigate dysbiosis and combat pathogens4,5.
The majority of the previously cited studies adopted a cross-sectional design, incorporating a relatively small number of patients. Yet, more longitudinal studies are needed to understand if this impairment is a cause or a consequence of DM and to elucidate how it might interfere with DM complications.