Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Similares em
SciELO
Similares em
SciELO
Compartilhar
Revista Portuguesa de Saúde Pública
versão impressa ISSN 0870-9025
Rev. Port. Sau. Pub. vol.34 no.3 Lisboa out. 2016
https://doi.org/10.1016/j.rpsp.2016.10.001
ORIGINAL ARTICLE
A large outbreak of Legionnaires’ Disease in an industrial town in Portugal
Um grande surto da Doença dos Legionários numa cidade industrial em Portugal
Francisco George a, Tara Shivaji a, Catia Sousa Pinto a, Luis Antonio Oliveira Serra a, João Valente a, Maria João Albuquerque a, Paula Cristina Olivença Vicêncio a, Ana San-Bento a, Paulo Diegues a, Paulo Jorge Nogueira a, Teresa Marques a, Helena Rebelo b, Filipa Costa b, Raquel Rodrigues b, Alexandra Nunes b, Vitor Borges b, João Paulo Gomes b, Daniel Sampaio b, Paula Barreiro b, Silvia Duarte b, Dina Carpinteiro b, Joana Mendonça b, Catarina Silva b, Luís Vieira b, Maria Joao Simões b, Paulo Gonçalves b, Baltazar Nunes b, Carlos Dias b, Jorge Machado b, Fernando Almeida b, Elsa A Goncalves c, Lucilia Carvalho d, Pedro Viterbo e, Dilia Jardim f, Nuno Lacasta f, Filomena Boavida f, Ana Perez f, Isabel Santana g, Paula Matias g, Nuno Banza g, Carlos Rabacal h
a Direção Geral da Saúde, Ministério da Saúde, Lisboa, Portugal
b Instituto Nacional de Saúde Dr. Ricardo Jorge, Ministério da Saúde, Lisboa, Portugal
c Centro Hospitalar de Lisboa Oriental, Ministério da Saúde, Lisboa, Portugal
d Centro de Estudos Anglísticos, Universidade de Lisboa, Ministério da Ciência, Tecnologia e Ensino Superior, Lisboa, Portugal
e Instituto Português do Mar e da Atmosfera, Ministério do Ambiente, Lisboa, Portugal
f Agência Portuguesa do Ambiente, Ministério do Ambiente, Amadora, Portugal
g Inspeção-Geral da Agricultura, do Mar, do Ambiente e do Ordenamento Territorial, Ministério do Ambiente, Ministério da Agricultura, Desenvolvimento Rural e Florestal e Ministério do Mar, Lisboa, Portugal
h Hospital de Vila Franca de Xira, Ministério da Saúde, Vila Franca de Xira, Portugal
Endereço para Correspondência
ABSTRACT
Background
We describe the investigation and control of an outbreak of Legionnaires’ disease in Portugal in October, November and December 2014.
Methods
Confirmed cases were individuals with pneumonia, laboratory evidence of Legionella pneumophila serogroup 1 and exposure, by residence, occupational or leisure to the affected municipalities. 49 possible sources were reduced to four potential sources, all industries with wet cooling system, following risk assessment. We geo-referenced cases’ residences and the location of cooling towers defining four study areas 10 km buffer centered on each cooling tower system. We compared the number of cases with expected numbers, calculated from the outbreak's attack rates applied to 2011 census population. Using Stones’ Test, we tested observed to expected ratios for decline in risk, with distance up to 10 km four directions. Isolates of Legionella pneumophila were compared using molecular methods.
Results
We identified 403 cases, 377 of which were confirmed, 14 patients died. Patients became ill between 14 October and 2 December. A NE wind and thermal inversion were recorded during the estimated period of exposure. Disease risk was highest in people living south west from all of the industries identified and decreased with distance (p < 0.001). 71 clinical isolates demonstrated an identical SBT profile to an isolate from a cooling tower. Whole genome sequencing identified an unusual L. pneumophila subsp. fraseri serogroup 1 as the outbreak causative strain, and confirmed isolates’ relatedness.
Conclusions
Industrial wet cooling systems, bacteria with enhanced survival characteristics and a combination of climatic conditions contributed to the second largest outbreak of Legionnaires’ disease recorded internationally.
Keywords: Legionnaires’ disease. Legionella pneumophila fraseri.
RESUMO
Contexto
Descrevemos a investigação epidemiológica e medidas de controlo de um surto de doença dos Legionários, ocorrido em Portugal em outubro, novembro e dezembro de 2014.
Métodos
A definição de caso englobou doentes com critérios clínicos de pneumonia aguda, com provas imagiológicas compatíveis e confirmação laboratorial para a identificação de Legionella pneumophila (L. pneumophila) serogrupo 1, para além do critério epidemiológico de exposição, quer por motivos de residência, ocupacional ou lazer nas freguesias suspeitas. Quarenta e nove possíveis fontes de infeção foram reduzidas a 4 potenciais fontes, após avaliação de risco, todas as indústrias com sistema de torres de arrefecimento. A georreferenciação por residência dos casos e localização de torres permitiu definir 4 áreas de investigação num perímetro de 10 km centrado em cada uma daquelas 4 torres. Comparou-se o número de casos observados com o número de casos esperados, calculados a partir de taxas de ataque do surto aplicadas à população. Usando o teste de Stones, testou-se a razão entre casos observados e casos esperados e declínio do risco em relação à distância de até 10 km em 4 direções. As amostras de L. pneumophila foram comparadas utilizando métodos moleculares.
Resultados
Foram identificados 403 casos, dos quais 377 foram confirmados, tendo ocorrido 14 óbitos. Os doentes apresentaram sintomas entre 14 de outubro e 2 de dezembro. Em termos meteorológicos, foram registados ventos NE e inversão térmica durante o período estimado de exposição. O risco de doença foi maior em pessoas que vivem a sudoeste de todas as indústrias identificadas, diminuindo com o aumento da distância (p < 0,001). Amostras de 71 dos casos clínicos demonstraram um perfil SBT idêntico às amostras isoladas a partir de uma torre de arrefecimento. A sequência de genoma de L. pneumophila fraseri serogrupo 1 pouco comum como a estirpe causadora do surto confirmou a relação das amostras isoladas.
Conclusões
Torres de arrefecimento industriais, agentes bacterianos com características mais desenvolvidas para elevada sobrevivência e uma rara combinação de condições climáticas, contribuíram para o segundo maior surto de doença dos Legionários registado na literatura.
Palavras-chave: Doença dos Legionários. Legionella pneumophila fraseri.
Introduction
On 7 November 2014, the Directorate-General of Health in Portugal was informed by a local hospital laboratory of 18 patients with Legionnaires’ disease, all admitted in the previous 24 h from Vila Franca de Xira, town on the outskirts of the capital city Lisbon. It rapidly became apparent that the numbers of cases were rising and cases were identified from across Portugal. A multidisciplinary taskforce investigated the outbreak, the results of their preliminary investigation led to the closure of industrial wet cooling systems. During the outbreak investigation, we presented a report that has been published as a rapid communication, providing preliminary data of the epidemiological, microbiological and environmental investigation.1 This article presents the final results of the outbreak investigation.
Legionnaires’ disease, first described during the 1976 Philadelphia American Legion conference2 is a bacterial pneumonic infection with Legionellae species. Most cases attributed to L. pneumophila sg 1.3 Disease characteristically develops 2–10 days after the inhalation of aerosolized bacteria by susceptible individuals. Risk factors include smoking, older individuals and those with chronic cardiorespiratory disease.4 Water, either in natural or artificial aquatic environments, is the reservoir for Legionellae species.5
From 2008 to 2012, between 88 and 140 cases were reported annually in Portugal of which the majority were sporadic community acquired cases.6 Large community outbreaks can result in significant morbidity and mortality in a short space of time,5 the largest to date was reported in 2001 in Murcia, Spain with 449 cases.7
Frequently identified as the source of large community outbreaks, industrial cooling tower systems source8 are able to disseminate contaminated aerosols over large distances.9 Meteorological factors, industrial operational conditions and inadequate maintenance are risk factors for Legionella outbreaks associated with towers.8,10
Methods
Definitions
A confirmed case of Legionnaires’ disease had radiologically confirmed pneumonia with symptom onset between 12 October ánd 02 December 2014, who lived or worked within 10 km of Vila Franca de Xira and had laboratory confirmation of infection. The laboratory criteria for a confirmed case included the isolation of Legionella spp. from respiratory secretions, the detection of L. pneumophila sg1 antigen in urine, a significant rise in specific antibody level to L. pneumophila sg 1 in paired serum samples. We defined a probable case differed only in the laboratory detection of either Legionella spp. nucleic acid in respiratory secretions or a single high level of specific antibody to L. pneumophila sg 1.
Identification of cases and assessment of exposure
Cases were identified by reviewing statutory laboratory and electronic clinical notifications from 1 October 2014. Staff from regional health authorities interviewed all confirmed and probable cases applying a standard Portuguese Legionnaires’ disease questionnaire which recorded demographic, epidemiological and clinical details. The residential address of each case was geocoded in Google Earth.
To identify deaths, we regularly cross-matched the outbreak database with the Portuguese real time information system for death certification (SICO), from November 2014 to March 2015.
Source identification
Possible sources of Legionella contamination, including industries with wet cooling systems, hospitals, malls and public recreation facilities were identified and, as a measure of precaution closed until environmental investigation. Operational and maintenance reports were checked and samples taken to screen for the presence Legionella spp., results led to the identification of potential sources. The locations of potential sources were mapped using visual pinpointing in Google Earth.
Microbiological, phenotypic and genotypic characterization
Detection of L. pneumophila sg 1 antigen was performed using specific urinary antigen tests in hospital laboratories. Respiratory and blood specimens were processed at the reference laboratory, the National Institute of Health Dr Ricardo Jorge. Environmental samples were obtained during inspections carried out by the General Inspectorate for Agriculture, Sea, Environment and Spatial Planning in accordance with Portuguese regulations, and processed by the at the reference laboratory, the National Institute of Health Dr Ricardo Jorge.
Clinical and environmental samples were analyzed using culture and/or molecular techniques. Clinical specimens were cultured on BCYE-based media and environmental samples were cultured in GVPC selective media. Isolates were identified by commercial latex-agglutination kits (Legionella latex Test, Oxoid, United Kingdom and Microgen Legionella, Microgen® Bioproducts,). Culture results for clinical samples were checked by real-time PCR targeted to the 16S and/or mip genes (L. pneumophila Real-TM, Sacace Biotechnologies, Italy; Argene Legio pneumo/Cc r-gene, bioMérieux, France). Isolates, or culture-negative/PCR positive specimens, were characterized by sequence-based typing (SBT) in accordance with the guidance of the European Working Group for Legionella Infections (EWGLI).11 The seven loci were subjected to Sanger sequencing by using BigDye v1.1 chemistry on a 3130XL Genetic Analyzer (Applied Biosystems). Whole genome sequencing (WGS) was performed for 10 clinical and one environmental sample on a MiSeq instrument (Illumina Inc., San Diego, USA) using using MiSeq V2 flow cells and 150 bp paired-end reads (depth of coverage >100-fold) to ascertain their similarity. Illumina reads were assembled using Velvet version 1.2.10.12
Phenotypic characterization of L. pneumophila sg1 used the Dresden panel of monoclonal antibodies (MAbs) consisting of six MAbs plus the MAb 3 of the International Panel obtained from the American Type Culture Collection (ATCC). Analysis followed an established algorithm, endorsed by EWGLI which can characterize L. pneumophila into one of nine subgroups; Knoxville, Philadelphia, France/Allentown, Benidorm, OLDA, Oxford, Heysham, Camperdown and Bellingham.13 The panel included antibodies to MAb 3/1 which is considered as a virulence marker due to high hydrophobicity of these strains.
Meteorological and air quality aspects
Data were obtained from the weather station in Vila Franca de Xira and the local air quality station (Alverca). Twice daily recordings of temperature and once daily recordings of humidity, air quality, wind speed and direction were collected for the period of 18 October–1 November 2014.
Statistical analysis
We conducted a source proximity analysis of four potential sources to test the hypothesis that risk of disease was inversely related to distance from the source. Using QGIS software, a buffer of 10 km radius further subdivided into concentric 1 km rings was placed on the towers of each industrial cooling system. The buffer and rings were then divided into four quadrants, North East, South East, South West and North West. We compared the number of cases with expected numbers. To calculate expected cases, age and sex specific rates for the outbreak were applied to the subsection population counts from the 2011 census. Subsections were categorized according to the distance and direction of their geographical centroid from each source. The expected number of cases for each band quadrant was calculated as the sum of the expected cases for the subsections whose centroids lay within it. Using Stones’ Test (ST), we tested observed to expected ratios for decline in risk, with distance up to 10 km in the North East, North West, South East and South West14 with 9999 Monte Carlo simulations and set the cut-off for statistical significance at 0.05.15
Back calculation models developed by Egan et al.16 were used to estimate the period of aerosolized release at the start of the outbreak. The models were repeated using cases notified up to March 2015. Statistical analysis was conducted using R statistical software.
Results
Outbreak description
By 2 December 2014, 403 cases of disease were identified of which 377 confirmed and 26 probable cases. Fourteen people died (case fatality 3.5%).
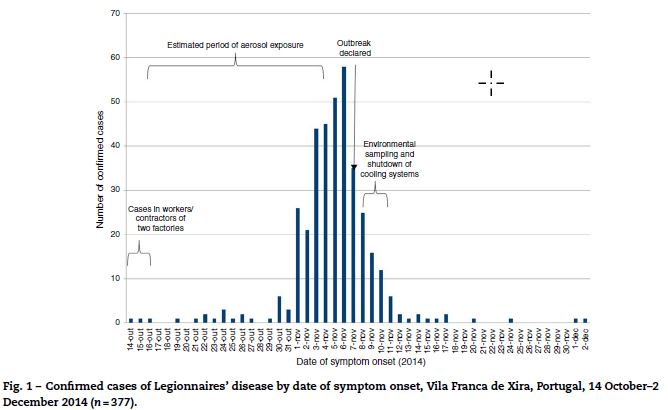
Confirmed case developed symptoms between 14 October and 2 December 2014. The number of cases peaked on 6 November and the outbreak was declared on 7 November (Fig. 1).
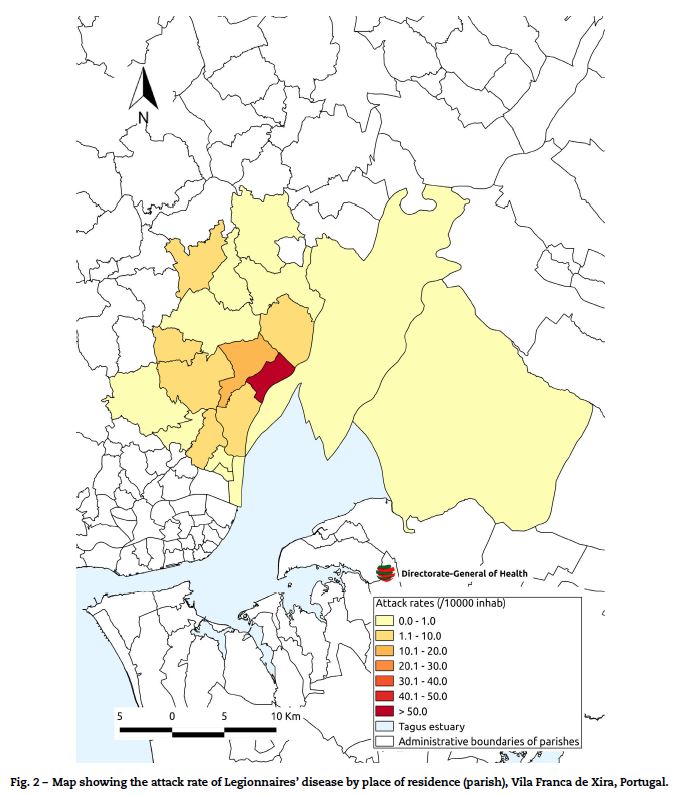
The highest attack rates were seen in the parish of Póvoa de Santa Iria/Forte de Casa (60 per 10,000 population) and dropped with increasing distance from this parish (Fig. 2).
There were 252 confirmed cases in men (66%). The median age was 59 years, the attack rate was higher in men and increased with age in both sexes (25.1 per 10,000 men aged 20–64 years versus 37.9 per 10,000 men aged over 65) and women (9.0 per 10,000 women aged 20–64 years versus 26.1 cases per 10,000 women aged over 65).
Two hundred and eight (55%) confirmed cases also reported increased disease susceptibility most commonly due to smoking and chronic cardio respiratory disease.
Source identification
Forty nine possible sources; domestic, public, commercial and industrial (5) were identified and reduced to four potential sources following environmental investigation. All four were industries with wet cooling systems.
Three out of the four potential sources reported suspected cases among employees, nine cases were confirmed.
Microbiological and phenotypic analysis and genetic characterization
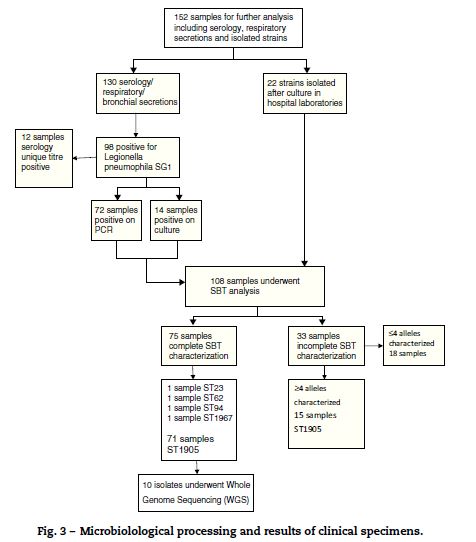
The reference laboratory received 95 environmental samples from 49 possible sources, of which, L. pneumophila positive cultures were isolated in 8. These samples originated from four industries with wet cooling systems and one commercial premises. Six of the 8 positive cultures were L. pneumophila sg 2–15 (75%). L. pneumophila sg 1 were isolated from two environmental samples taken from the same tower of one industrial system, recorded concentrations were 1.42 × 106 ufc/l on 8 November and 421 ufc/l on 10 November. Sequence-based typing of these two environmental isolates identified the novel genotype ST1905.
Laboratory confirmation of cases was done by; urinary antigen testing for L. pneumophila sg 1 (92%); seroconversion (2%); PCR (3%) and unique titer to L. pneumophila (3%). 152 clinical specimens were sent to the reference laboratory. Of these, 71 revealed the presence of L. pneumophila sg 1 with identical ST1905 SBT profile.
37 isolates of L. pneumophila sg 1 underwent phenotypic characterization, 35 isolates from clinical samples and two from environmental samples. All 37 isolates presented the same phenotypic characteristic; subtype France/Allentown and possessed the virulence-associated epitope recognized by monoclonal antibody MAb 3/1 (Fig. 3).
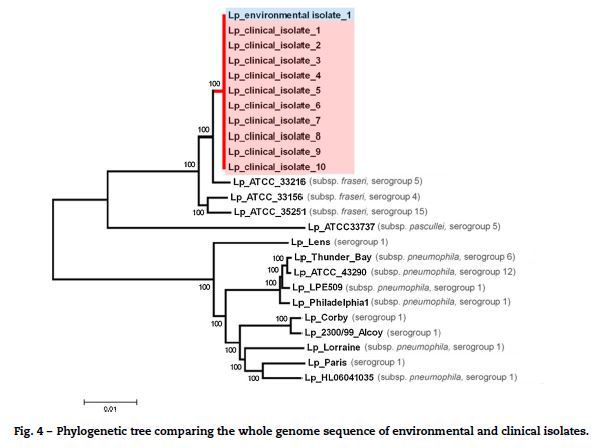
To confirm the genetic relatedness of ST1905 isolates, 10 out of the 71 clinical isolates and one environmental positive isolate obtained from the industrial cooling tower were nominally selected for whole genome sequencing. We were able to use about 99.8% of each draft sequence and found no nucleotide differences within the compared 3.47 Mb of the genome. Phylogenetic analysis involving multiple L. pneumophila isolates from across the world revealed that the ST1905 cluster clearly diverged from the branch enrolling the most studied L. pneumophila strains (serogroups 1, 6 and 12)17–19 and was more closely related with L. pneumophila subsp. fraseri strains (serogroup 4, 5 and 15).20 WGS confirmation of the ST1905 allelic profile highlighted a bias associated with the in silico extraction of the allele sequence for the mompS locus. The studied strain displayed non-matching mompS copies, which hampered a proper ST attribution if SBT was exclusively determined in silico (Fig. 4).
Meteorological, climate, and air quality results
Temperatures in excess of the monthly average were recorded from 18 October to 1 November (>20 °C), with a persistent nocturnal thermal inversion associated to a predominant NE and NNE wind with 2–3 km/h speed. The last week of October had more than 80% relative humidity, with periods exceeding 90%. October 2014 was the warmest October in mainland Portugal since 1931, including an 11 days heat wave during the 17–27 October period. From 19 October to 1 November, three times the recommended level of particulate matter (PM10) and the highest weekly values in 2014, was recorded in the local air quality station, attributed to a strong episode of Sahara dust transported to mainland Portugal.
Period of exposure assessment
The best fitting back-calculation models suggested a logistic growth of the environmental exposure, with greatest intensity between 16 October and 4 November (Fig. 1).
Source proximity analysis results
When the source proximity analysis was conducted, there were 365 confirmed cases, of which 328 (90%) were geo-referenced to within 10 km of one of the four potential sources (Fig. 5).
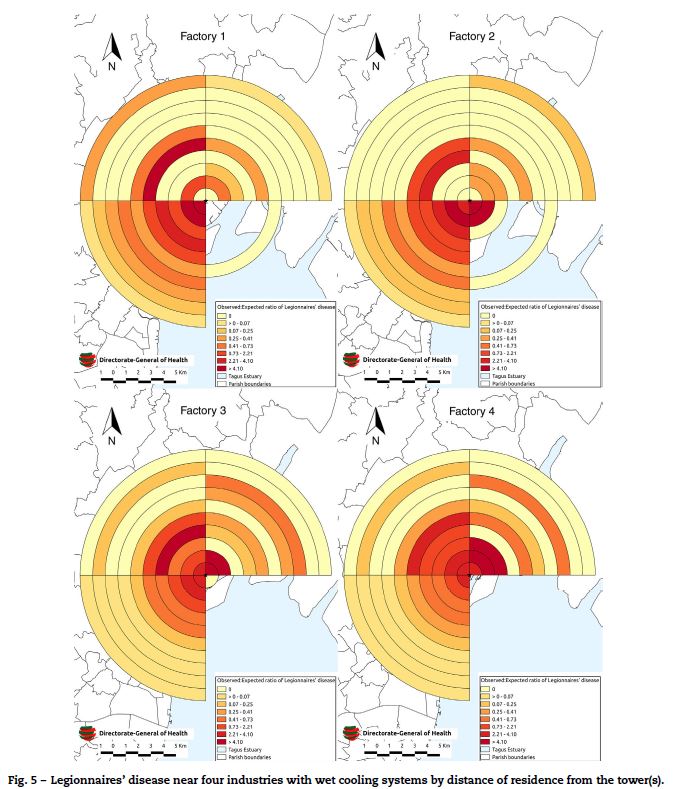
All four potential sources demonstrated a significant decline in disease risk with increasing distance (clustering) in the SW Quadrant. The highest clustering (ST157, p < 0.001) was observed in the SW quadrant of industry 1. Clustering was also observed with industry 2 (ST 117, p < 0.001), industry 3 (ST 126, p < 0.001) and industry 4 (ST 66.1, p < 0.001). The risk of disease fell consistently with increasing distance from the source with no evidence of peaks or troughs in this quadrant. No clustering was observed in the SE quadrants of any industry, an area that largely comprised of the river. Industry 1 and 2 demonstrated a lower degree of clustering in the NE quadrant (ST 5, p = 0.01 and ST 4.2, p = 0.01 respectively). Clustering was also demonstrated in the NW quadrant of industry 1 (ST 3.9, p = 0.01).
Discussion
We identified four industries with wet cooling system as the most likely sources of the second largest outbreak of Legionnaires’ disease recorded with 403 cases and 14 deaths. Elevated concentrations of L. pneumophila sg 1 ST1905 demonstrating the virulent MAb 3–1 subgroup were identified in the towers of one industry. Using SBT and WGS, we were able to confirm the genetic relatedness of the outbreak-associated clinical and environmental strains. The only positive isolates for L. pneumophila sg 1, were both obtained from the cooling tower of one industry. Similar concentrations of L. pneumophila have been isolated in other cooling towers associated with outbreaks.5 It is probable, though unproven that cross contamination of the four closely located towers contributed to the magnitude of the outbreak and has previously been described.21
The unseasonably warm temperatures recorded during October 2014 supported the proliferation of Legionellae species in cooling tower systems.22 The high PM10 concentrations might have acted as cloud concentration nuclei facilitating aerosol formation. The distribution and large number of cases are accounted for by the NE wind, high relative humidity and thermal inversion observed during the estimated release period in addition to the longevity23 and heightened virulence24 of a bacterium with epitope MAb3/1.
The outbreak occurred in a densely populated area close to the capital city of Portugal containing heavily used commuter routes from suburban areas. The actual numbers of people exposed was likely to have been much higher than the resident population. Standard diagnostic tests may not detect all cases.3 The figures presented do not contain cases of Pontiac fever or suspected cases who tested negative and therefore probably underestimates the morbidity experienced by the community and impact on local health services. In contrast due to the presence of electronic death notification in Portugal, it was possible to ensure that information on whether a case had died was complete. The observed case fatality (3.5% of all cases) was lower than the 6% usually observed in outbreaks associated with cooling towers and systems.8 The outbreak causative strain phylogenetically clustered apart from the most studied outbreak-associated L. pneumophila sg 1 strains (Lp Philadelphia-1, Lp Paris, Lp Lens, Lp Corby, and Lp 2300/99 Alcoy). It was found to be closely related to L. pneumophila subsp. fraseri strains from serogroups other than sg 1. The studied strain harbors an exclusive ∼38 kb genomic region compared with the next most phylogenetically related strain. This region was found to be intact and highly similar (BLASTn, cover 100%, identity 99%, E-value 0.0) only in one strain (ATCC 33761 = DSM 21215) of L. oakridgensis, a species that rarely causes Legionnaires’ disease.25 We would explain the low case fatality observed in terms of early diagnosis and appropriate and timely treatment, whether the unique traits of the outbreak-associated strain underlie important characteristics of lethality remains under study.
The source proximity analysis estimated expected numbers of cases from the resident population, which likely led to an underestimation of exposure. We reasoned that residential information was collected systematically, unlike displacement history, also conditions for the horizontal propagation of aerosols were more pronounced at night time when most residents would have been in their homes. A more robust approach would have to perform the analysis as part of a case control study, which could also include exposures in non residents. Although initially discussed, we did not proceed with a case–control study due to resource and time constraints. The close geographical proximity of the towers meant that there was overlap of some of the study areas rendering it possible, but unlikely that a closely located source could lead to presence of a distance decline relationship for another source. Comparing the geospatial distribution of cases with plume modeling of aerosol could be of value to assess the impact of closely located sources on each other.
This outbreak was the second largest outbreak of Legionnaires’ disease recorded to date and adds to the body of evidence regarding the impact of climatic conditions and bacterial phenotypes in cooling tower associated community outbreaks of Legionnaires’ disease.
References
1. Shivaji T., Sousa C., San-Bento A., Oliveira L.A., Valente J., Machado J., et al. A large community outbreak of Legionnaires’ disease in Vila Franca de Xira, Portugal, October to November 2014. Euro Surveill. 2014;19.
2. Fraser D.W., Tsai T.R., Orenstein W., Parkin W.E., Beecham H.J., Sharrar R.G., et al. Legionnaires’ disease: Description of an epidemic of pneumonia. N Engl J Med. 1977;297:1189-97.
3. Fields B.S., Benson R.F., Besser R.E. Legionella and Legionnaires’ disease: 25 years of investigation. Clin Microbiol Rev. 2002;15:506-26.
4. Phin N., Parry-Ford F., Harrison T., Stagg H.R., Zhang N., Kartik K., et al. Epidemiology and clinical management of Legionnaires’ disease. Lancet Infect Dis. 2014;14:1011-21.
5. Legionella and the prevention of legionellosis. Geneva: WHO, (2007) pp. 40. [ Links ]
6. Annual epidemiological report 2014, respiratory tract infections. Stockholm: ECDC, (2014) pp. 17. [ Links ]
7. García-Fulgueiras A., Navarro C., Fenoll D., García J., González-Diego P., Jiménez-Buñuales T., et al. Legionnaires’ disease outbreak in Murcia, Spain. Emerg Infect Dis. 2003;9:915-21.
8. Walser S.M., Gerstner D.G., Brenner B., Höller C., Liebl B., Herr C.E. Assessing the environmental health relevance of cooling towers: A systematic review of legionellosis outbreaks. Int J Hyg Environ Health. 2014;217:145-54. [ Links ]
9. Nguyen T.M.N., Ilef D., Jarraud S., Rouil L., Campese C., Che D., et al. A community-wide outbreak of legionnaires disease linked to industrial cooling towers: How far can contaminated aerosols spread?. J Infect Dis. 2006;193:102-11. [ Links ]
10. Mouchtouri V.A., Goutziana G., Kremastinou J., Hadjichristodoulou C. Legionella species colonization in cooling towers: Risk factors and assessment of control measures. Am J Infect Control. 2010;38:50-5. [ Links ]
11. Gaia V., Fry N.K., Afshar B., Lück P.C., Meugnier H., Etienne J., et al. Consensus sequence-based scheme for epidemiological typing of clinical and environmental isolates of Legionella pneumophila. J Clin Microbiol. 2005;43:2047-205. [ Links ]
12. Zerbino D.R., Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821-9. [ Links ]
13. Helbig J.H., Sverker B., Pastoris M.C., Etienne J., Gaia V., Lauwers S., et al. Pan-European study on culture-proven Legionnaires’ disease: Distribution of Legionella pneumophila serogroups and monoclonal subgroups. Eur J Clin Microbiol Infect Dis. 2002;21:710-6.
14. Bull M., Hall I.M., Leach S., Robesyn E. The application of geographic information systems and spatial data during Legionnaires disease outbreak responses. Euro Surveill. 2012;17:20331. [ Links ]
15. Tango T. Statistical methods for disease clustering. New York, NY: Springer Science & Business Media, (2010) pp. 191. [ Links ]
16. Egan J.R., Hall I.M., Lemon D.J., Leach S. Modeling Legionnaires’ disease outbreaks: Estimating the timing of an aerosolized release using symptom-onset dates. Epidemiology. 2011;22:188-98.
17. Khan M.A., Knox N., Prashar A., Alexander D., Abdel-Nour M., Duncan C., et al. Comparative genomics reveal that host-innate immune responses influence the clinical prevalence of Legionella pneumophila serogroups. PLoS ONE. 2013;8:e67298. [ Links ]
18. Gomez-Valero L., Rusniok C., Jarraud S., Vacherie B., Rouy Z., Barbe V., et al. Extensive recombination events and horizontal gene transfer shaped the Legionella pneumophila genomes. BMC Genomics. 2011;12:536. [ Links ]
19. Ma J., He Y., Hu B., Luo Z.Q. Genome sequence of an environmental isolate of the bacterial pathogen Legionella pneumophila. Genome Announc. 2013;1:e00320-13. [ Links ]
20. Ko K.S., Lee H.K., Park M.Y., Park M.S., Lee K.H., Woo S.Y., et al. Population genetic structure of Legionella pneumophila inferred from RNA polymerase gene (rpoB) and DotA gene (dotA) sequences. J Bacteriol. 2002;184:2123-30. [ Links ]
21. Kool J.L., Buchholz U., Peterson C., Brown E.W., Benson R.F., Pruckler J.M., et al. Strengths and limitations of molecular subtyping in a community outbreak of Legionnaires’ disease. Epidemiol Infect. 2000;125:599-608.
22. Bentham R.H., Broadbent C.R. A model for autumn outbreaks of Legionnaires’ disease associated with cooling towers, linked to system operation and size. Epidemiol Infect. 1993;111:287-95.
23. Ditommaso S., Giacomuzzi M., Rivera S.R., Raso R., Ferrero P., Zotti C.M. Virulence of Legionella pneumophila strains isolated from hospital water system and healthcare-associated Legionnaires’ disease in Northern Italy between 2004 and 2009. BMC Infect Dis. 2014;14:483.
24. Helbig J.H., Lück P.C., Knirel Y.A., Witzleb W., Zähringer U. Molecular characterization of a virulence-associated epitope on the lipopolysaccharide of Legionella pneumophila serogroup 1. Epidemiol Infect. 1995;115:71-8. [ Links ]
25. Brzuszkiewicz E., Schulz T., Rydzewski K., Daniel R., Gillmaier N., Dittmann C., et al. Legionella oakridgensis ATCC 33761 genome sequence and phenotypic characterization reveals its replication capacity in amoebae. Int J Med Microbiol. 2013;303:514-28. [ Links ]
Conflicts of interest
The authors have no conflicts of interest to declare.
Acknowledgements
Organizations and individuals involved in the Task Force for the Legionnaires’ Disease outbreak in Vila Franca de Xira; staff from Hospital de Vila Franca de Xira, Hospital CUF Descobertas, Centro Hospitalar de Lisboa Central, Centro Hospitalar de Lisboa Norte; Centro Hospitalar de Lisboa Ocidental; staff of the health authorities of Lisbon and Tagus Valley, central Portugal, north Portugal and the Algarve. Dr. Christian Lucke, Tecnische Universitat Dresden, for supplying the MAbs reagents. Marion Muelhen, and Paula Vasconcelos for critical review of the manuscript. Emmanuel Robesyn for technical advice on geospatial analysis techniques in Legionnaires’ disease.
Received October 13, 2016
Accepted October 14, 2016
Corresponding author