Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Similares em
SciELO
Similares em
SciELO
Compartilhar
Revista Portuguesa de Pneumologia
versão impressa ISSN 0873-2159
Rev Port Pneumol v.15 n.5 Lisboa out. 2009
Genotypic analysis of Mycobacterium tuberculosis isolates from a Lisbon hospital in Portugal
João Perdigão 1
Catarina Milho 2
Lurdes Carrilho 3
Laura Brum 4
Isabel Portugal 5
Abstract
Portugal has one of the highest tuberculosis notification rates of the European Union with Lisbon Health Region having an incidence rate well above the national average. The present study analyses the transmission, drug susceptibility and characteristics of a study population from a Central Lisbons Hospital.
One hundred and thirty-two Mycobacterium tuberculosis clinical isolates were previously tested for drug susceptibility to first-line drugs. The multidrug (MDR) resistance rate was found to be 3.0%, while 13.6% of the isolates were resistant to one or more first-line drugs. HIV serology was available for 98 patients, 26 (26.5%) were positive. Genotyping was performed by MIRU-VNTR and 53 (40,2%) out of the 132 isolates were found to be distributed through 17 MIRU -VNTR clusters of two or more isolates.
Lisboa strains accounted for 25.8% of all strains. We conclude that transmission of resistant and susceptible Mycobacterium tuberculosis strains is occurring, with special concern for Lisboa strains.
Key-words: Lisboa family, tuberculosis, multidrug resistance, MIRU.
Análise genotípica de isolados de Mycobacterium tuberculosis de um hospital em Lisboa, Portugal
Resumo
Portugal apresenta uma das taxas de incidência de tuberculose mais altas da União Europeia, tendo a Região de Saúde de Lisboa uma taxa de incidência muito acima da média nacional. O presente estudo analisa a transmissão, susceptibilidade aos antibacilares e características de uma população de estudo de um hospital central de Lisboa. Cento e trinta e dois isolados clínicos de Mycobacterium tuberculosis foram previamente testados quanto à susceptibilidade aos antibacilares de primeira linha. A taxa de multirresistência encontrada foi de 3,0%, enquanto 13,6% dos isolados eram resistentes a um ou mais antibacilares de primeira linha. A serologia para o VIH estava disponível para 98 doentes, 26 (26,5%) eram positivos. Os isolados foram genotipados por MIRU-VNTR e 53 (40,2%) dos 132 isolados encontravam-se distribuídos por 17 clusters MIRU-VNTR diferentes de dois ou mais isolados. De todos os isolados analisados, 25,8% pertenciam à família Lisboa. Concluímos que a transmissão de estirpes de Mycobacterium tuberculosis, resistentes e susceptíveis, está a ocorrer, com especial preocupação para as estirpes Lisboa.
Palavras-chave: Família Lisboa, tuberculose, multirresistência, MIRU.
Introduction
The tuberculosis (TB) situation in Portugal is one of the most serious of the European Union (EU). In 2006, Portuguese health authorities have reported a TB incidence rate of 32.4 cases per 100 000 inhabitants.
Portugal was only surpassed by Bulgaria, Estonia, Latvia, Lithuania and Romania1. The distribution of the TB incidence across the Portuguese territory is not equal. The district of the capital city, Lisbon and the district of the second largest city, Oporto, had in 2006 TB incidence rates of 36.9 and 45.4 cases per 100 000 inhabitants, respectively2.
This is an incidence rate well above the national average. In addition, there is the problem of drug resistance in Portugal. In fact, in 2006, Portugal reported a multidrug resistance (MDR) rate of 1.5%, a number that was shown by some of us to be underestimated3.
In Lisbon, approximately three quarters of all tuberculosis cases occur among three major risk groups: human immunodeficiency virus (HIV) infected, drug users and immigrants4.
The same region was shown, in 2005, to have several high-risk areas (>3 cases and >100 cases per 100 000 inhabitants).
The drug resistant strains causing MDR–TB in Lisbon region have been extensively studied in the last 15 years. This has allowed the identification of a family of strains, designated Lisboa family, responsible for the majority of MDR-TB cases in the region3,5,6.
The use of molecular epidemiology techniques for genotyping mycobacterial strains has contributed to a better knowledge of TB transmission dynamics, to the indentification of epidemiological links between patients and, to TB control in a given area7-11.
For many years, Restriction Fragment Length Polymorphism with hybridization of IS6110 probe (RFLP -IS6110), used as a molecular epidemiological tool, has proven worthy in the discrimination of mycobacterial strains due to its stability and discriminative power7,12. Nevertheless in the last years this time-consuming technique has been gradually substituted for the characterization of Mycobacterial Interspersed Repetitive Units – Variable Tandem Repeats (MIRU -VNTR)8,13,14. This new tool has progressively been improved to give a discriminative power equal to that of RFLP- IS611015,16.
In the present study we have genotyped resistant and susceptible Mycobacterium tuberculosis (M. tuberculosis) clinical isolates from a specific hospital by MIRU-VNTR, in order to better understand the transmissibility and clonality of M. tuberculosis strains circulating in Lisbon area.
Materials and methods
Study population
TB patients from Hospital Pulido Valente in Lisbon were studied during the year of 2006. This hospital is one of the main hospitals in Lisbon performing TB diagnosis and treatment, and in 2006, 211 TB patients were received and diagnosed with positive culture. From these, 132 M. tuberculosis clinical isolates were available for further characterization in the reference laboratory of the National Institute of Health.
Clinical isolates and DNA extraction
The 132 M. tuberculosis clinical isolates, each corresponding to one patient, were isolated and grown on BACTEC® 960 MGIT® (Becton -Dickinson®, Franklin Lakes, USA) and Lowenstein-Jensen slants. Identification was performed with Accuprobe® (Gen-Probe®, San Diego, USA).
Crude DNA extracts were produced by heating a loopful of bacterial growth in TE Buffer solution to 100.ºC for 10 minutes, followed by sonic bath incubation for 15 minutes. Finally, the mixture was centrifuged at maximum speed for 15 minutes and the supernatant used in the subsequent molecular biology reactions.
Drug susceptibility testing
Isolates were tested for first-line drug susceptibility using the BACTEC 960 MGIT (Becton-Dickinson®, Franklin Lakes, USA) methodology for isoniazid (0.1 μg/ml), rifampicin (1.0 μg/ml), streptomycin (1.0 μg/ml), ethambutol (5.0 μg/ml) and pyrazinamide (100 μg/ml), according to the manufacturer's instructions.
Isolates resistant to at least isoniazid and rifampicin were considered as MDR-TB.
MIRU-VNTR genotyping
MIRU-VNTR genotyping was performed for each isolate by multiplex PCR amplification of 12 MIRU-VNTR lociusing Hot-StarTaq® DNA polymerase (Quiagen®, Hilden, Germany), as previously described by Supply et al (2001).
Clustering analysis
Isolates were clustered accordingly to their MIRU-VNTR profile using BioNumerics-Latem, Belgium). Pearson coefficient was used to calculate the similarity matrix and clustering was performed by the unweighted pair group method with arithmetic mean (UPGMA). A cluster was defined as a group of two or more isolates with equal MIRU-VNTR profiles. Rate of genetic diversity was calculated by dividing the number of different MIRU-VNTR profiles by the total number of genotyped strains. The rate of recent transmission was calculated by division of the number of clustered strains, minus the number of clusters, by the total number of genotyped strains17.
Statistical association analysis
Analysis of resistance and HIV associated with clustering phenomenon was undertaken by use of the Fishers exact test, calculating p-values, odds ratios (OR) and confidence interval (CI) of 95%.
Geographical mapping of patients
The geographical distribution of patients was performed with ArcGIS® 9.1 (ESRI®, Redlands, USA) using maps and postal code information available at http://www.ctt.pt.
Results
Study population characterization
In this study we analyzed 132 M. tuberculosis clinical isolates, each one corresponding to a different patient. Of these, 105 patients were male (79,5%) and 27 were female (20,5%).
The patients age ranged between 16 and 95 years, with a median age of 43 years old. HIV serology data was available for 98 patients, of which 26 (26.5%) were HIV positive. HIV co-infection was higher in patients ranging between 30-54 years old. In these age groups we also found high frequency of TB cases (Fig. 1).
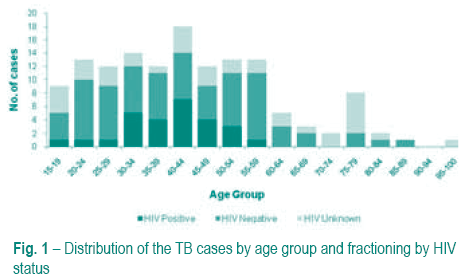
Drug resistance
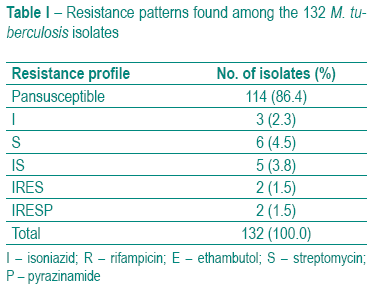
All 132 isolates were tested for resistance to all first-line drugs. Eighteen (13.6%) isolates, distributed through five different resistance patterns, were resistant to one or more antibacillary drugs (Table I). MDR-TB accounted for 3% (4/132) of all isolates tested.
MIRU-VNTR cluster analysis
All 132 isolates were characterized by the MIRU-VNTR technique as described by Supply et al (2001). Seventeen clusters containing 53 (40.2%) out of the 132 isolates were found (Fig. 2). Nine clusters, containing a total of 34 (25.8%) isolates belonged to a family of strains designated Lisboa family3,5.
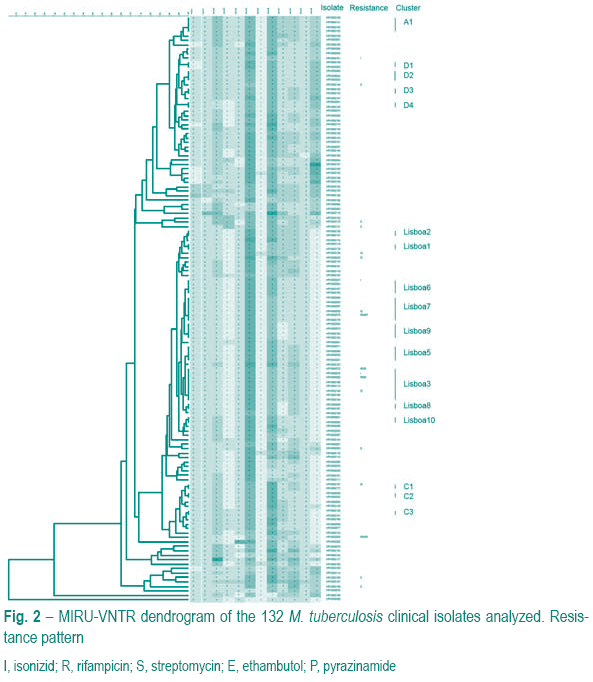
Three of the four MDR-TB cases belonged to Lisboa family. Fourteen Lisboa strains were not included in any cluster. Although we had a high frequency of HIV isolates in cluster, HIV co-infection was not statistically correlated with being in cluster (OR=2.391; CI95%=0.9410-6.075 p=0.0852). Drug resistance was also not statistically associated with clustering in this study (OR=1.227; CI95%=0.4499-3.344 p=0.7971).
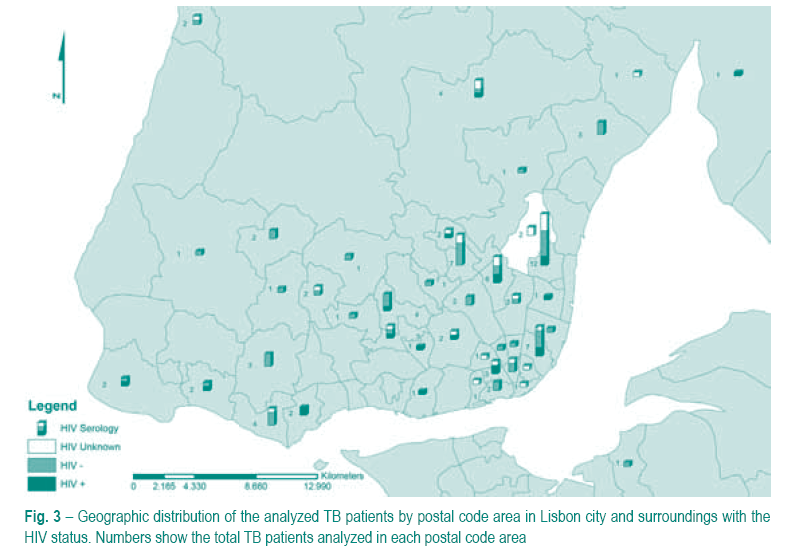
Geographical distribution of patients
We examined the geographical distribution of the 132 TB patients. The patients were distributed by the postal code area of their residence. This information was available for 118 patients; the remaining 14 were excluded from this analysis. Given the fact that Hospital Pulido Valente primarily serves the Lisbon region, we concentrated this analysis in this same region and surroundings (Fig. 3). Four postal code areas were found to have more than 5 cases. In fact, one of the postal code area had 12 of the 132 analyzed cases.
Discussion
In this study we have analyzed 132 M. tuberculosis clinical isolates originating from a single hospital in Lisbon Health region. The patients had a median age of 43 years old, which is in agreement with the Portuguese health authorities report. When we analyzed the HIV status of all patients we have observed that the TB high incidence age groups also had a high prevalence of HIV infected patients.
All clinical isolates were routinely subjected to drug susceptibility testing for first–line drugs. The drug resistance frequency was higher in this population than the average reported for the whole country1, with special attention for the MDR isolates which accounts for 3% of the study population.
Lisbon health region has been in the last 10-15 years a problematic region concerning drug resistance and MDR3,5,6. The application of molecular epidemiological methods in the characterization of Lisbon strains in the 90s unveiled a MDR-TB outbreak occurring mainly in the HIV-infected population5,6.
More recently, the same strains, belonging to the designated Lisboa family, were shown to significantly continue to circulate, evolving from MDR to extensive drug resistance (XDR)3.
All isolates in this study were genotyped by MIRU-VNTR. Seventeen clusters, containing 40.2% of all isolates, were found. We estimate that 27% of the studied cases are probably due to recent transmission. However the true dimension of TB cases due to phenomena of recent transmission is probably higher than our study estimates since only one hospital was analyzed. This hospital receives about 20% of the total TB cases in Lisbon region.
A considerable proportion of the isolates (25.8%) belonged to Lisboa family, including three of the MDR-TB strains. Nine clusters belonging to this family were found, including more than 64% of the clustered isolates, suggesting a higher relative transmissibility of these strains. This transmissibility and persistence in the community may be due to several factors, namely, the fact that the original HIV-infected niche may act as a reservoir more difficult to act on and, MDR may increase the persistence and the infectious period in the host. Therefore, this would provide more time for transmission to occur.
The most prevalent cluster, containing 8 (6.1%) of all isolates, is designated Lisboa3 and has been identified consecutively each year, across several health institutions, maintaining its predominance among resistant and susceptible strains. Belonging to this cluster we found not only drug –resistant strains but also pan-susceptible strains that may represent ancestor clones of the current MDR strains. These, have evolved by acquiring resistance to the different antibacillary drugs followed by clonal expansion.
Clusters containing pansusceptible isolates only, have been detected and represent minor transmission events of these type of strains. Nevertheless, in Lisboa 9 cluster, 3 of 4 isolates are also HIV-infected, which may pose a threat as it may favour the acquisition of resistance or even MDR of a strain that is already circulating at an apparently high frequency. In cluster A1 the opposite situation can be found, transmission of a susceptible strain has occurred among immunocompetent individuals only. Special attention must be paid in order to contain this strain before it reaches immunosupressed individuals, causing then a more serious public health problem.
We looked at the geographical distribution of the analyzed patients in an attempt to identify geographical hotspots were TB transmission may be occurring. It was found that the cases here analyzed had not an equal distribution across their respective residence areas, with some of these areas having a significantly higher number of cases than others. In addition, patients living on the same postal code did not, in most cases, belonged to the same genetic cluster (data not shown). Although there is the possibility that some of the patients addresses may not correspond to their real residence, our data suggests that active TB transmission might be occurring mainly outside the area of residence.
Comparative genomic studies of M. tuberculosis clinical isolates have identified regions of difference and single nucleotide polymorphisms among different lineages or families of strains18-22. These genomic differences may account for differentiated or even increased virulence, faster acquisition of resistance or, higher transmissibility19,23-25. A more thorough genomic and physiological characterization of Lisboa strains is ongoing and will provide more clues to its virulence and ability to spread.
Throughout the world special attention must be paid to local prevalent families of strains. One of the most recent examples is the KZN family of strains that in 10 years evolved from pansusceptibility to XDR. These strains caused a XDR-TB outbreak associated with high mortality rates in South Africa26,27. Lisboa strains have also evolved to XDR and may cause a XDR-TB outbreak if they remain uncontrolled. Given the difficulty, and sometimes the impossibility of treatment of XDR-TB, such an outbreak would constitute a very serious public health problem.
Conclusion
In conclusion, no significant TB outbreak was detected among the patients attending this studied hospital, although a considerable proportion of the isolates belonged to Lisboa family and approximately 27% of all cases were probably due to recent transmission.
Although we only found four MDR-TB strains, these poses a serious public health problem and measures are necessary to contain them. Susceptible Lisboa strains should also be monitored, especially those infecting HIV positive individuals, to prevent the acquisition of resistance.
Acknowledgments
This work was supported by Fundação para a Ciência e Tecnologia (FCT grant POCTI/ESP/42941/2001). The authors wish to thank to the working group participating in the VigLab surveillance system for data collection and processing. The authors are also grateful to the personnel from the Secção de Micobactérias do Serviço de Patologia Clínica do Hospital Pulido Valente and Unidade de Micobactérias do Departamento de Doenças Infecciosas do Instituto Nacional de Saúde Dr. Ricardo Jorge, whose high quality of service and cooperation have made this work possible.
Bibliography
1. EuroTB. Surveillance of tuberculosis cases in Europe. Report on tuberculosis cases notified in 2006. Saint-Maurice, France: Institut de veille sanitaire, 2008.
2. DGS. Programa Nacional de Luta contra a Tuberculose. Tuberculose: Ponto da Situação em Portugal em 2006, dados preliminares em Março 2007. Lisbon, Portugal: Direcção-Geral de Saúde, 2007.
3. Perdigão J, et al. Multidrug-resistant tuberculosis in Lisbon, Portugal: A Molecular Epidemiological Perspective. Microb Drug Resist 2008;14(2):133-143. [ Links ]
4. DGS. Programa Nacional de Luta contra a Tuberculose. Ponto da situação epidemiológica e indicadores de desempenho, Ano de 2003. Lisbon, Portugal: Direcção-Geral de Saúde, 2005.
5. Portugal I, et al. Outbreak of multiple drug–resistant tuberculosis in Lisbon: detection by restriction fragment length polymorphism analysis. Int J Tuberc Lung Dis 1999;3(3):207-213. [ Links ]
6. Portugal I, Maia S, Moniz-Pereira J. Discrimination of multidrug-resistant Mycobacterium tuberculosis IS6110 fingerprint subclusters by rpoB gene mutation analysis. J Clin Microbiol 1999. [ Links ]
7. Alland D, et al. Transmission of tuberculosis in New York City. An analysis by DNA fingerprinting and conventional epidemiologic methods. N Engl J Med 1994;330(24):1710-1716.
8. Barnes PF, Cave MD. Molecular epidemiology of tuberculosis. N Engl J Med 2003;349(12):1149-1156.
9. Small PM, et al. The epidemiology of tuberculosis in San Francisco. A population-based study using conventional and molecular methods. N Engl J Med 1994; 330(24):1703-1709.
10. Weis SE, et al. Transmission dynamics of tuberculosis in Tarrant county, Texas. Am J Respir Crit Care Med 2002;166(1):36-42.
11. Golub JE, et al. Transmission of Mycobacterium tuberculosis through casual contact with an infectious case. Arch Intern Med 2001;161(18):2254-2258.
12. van Soolingen D, et al. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol 1991; 29(11):2578-2586.
13. Blackwood KS, Wolfe JN, Kabani AM. Application of mycobacterial interspersed repetitive unit typing to Manitoba tuberculosis cases: can restriction fragment length polymorphism be forgotten? J Clin Microbiol 2004;42(11):5001-5006.
14. Supply P, et al. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J Clin Microbiol 2001;39(10):3563-3571.
15. Supply P, et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol 2006; 44(12):4498-4510.
16. van Deutekom H, et al. Molecular typing of Mycobacterium tuberculosis by mycobacterial interspersed repetitive unit-variable-number tandem repeat analysis, a more accurate method for identifying epidemiological links between patients with tuberculosis. J Clin Microbiol 2004;43(9):4473-4479.
17. Murray M, Alland D. Methodological problems in the molecular epidemiology of tuberculosis. Am J Epidemiol 2002;155(6):565-571.
18. Olano J, et al. Mutations in DNA repair genes are associated with the Haarlem lineage of Mycobacterium tuberculosis independently of their antibiotic resistance. Tuberculosis 2007;87(6):502-508.
19. Tsolaki AG, et al. Genomic deletions classify the Beijing/W strains as a distinct genetic lineage of Mycobacterium tuberculosis. J Clin Microbiol 2005; 43(7):3185-3191.
20. Tsolaki AG, et al. Functional and evolutionary genomics of Mycobacterium tuberculosis: insights from genomic deletions in 100 strains. Proc Natl Acad Sci USA 2004;101(14):4865-4870.
21. Brosch R, et al. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc Natl Acad Sci USA 2002;99(6):3684-3689.
22. Filliol I, et al. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J Bacteriol 2006;188(2):759-772.
23. Lopez B, et al. A marked difference in pathogenesis and immune response induced by different Mycobacterium tuberculosis genotypes. Clin Exp Immunol 2003;133(1):30-37.
24. Newton SM, et al. A deletion defining a common Asian lineage of Mycobacterium tuberculosis associates with immune subversion. Proc Natl Acad Sci USA 2006; 103(42):15 594-15 598.
25. Rad ME, et al. Mutations in putative mutator genes of Mycobacterium tuberculosis strains of the W–Beijing family. Emerg Infect Dis 2003; 9(7):838-45.
26. Gandhi NR, et al. Extensively drug-resistant tuberculosis as a cause of death in patients co-infected with tuberculosis and HIV in a rural area of South Africa. Lancet 2006; 368(9547):1575-1580.
27. Davies GR, et al. Emergence of multidrug–resistant tuberculosis in a community-based directly observed treatment programme in rural South Africa. Int J Tuberc Lung Dis 1999; 3(9):799-804.
1 Licenciado em biologia microbiana e genética, mestre em biologia molecular e genética, Aluno de doutoramento em microbiologia/BsC, Microbiology and Genetics. MsC Molecular Biology and Genetics. PhD Microbiology (partway)
2 Licenciada em microbiologia/BsC, Microbiology Molecular Centro de Patogénese Molecular, URIA, Faculdade de Farmácia da Universidade de Lisboa, Portugal/Molecular Pathogenesis Centre, URIA, School of Pharmacy, Universidade de Lisboa, Portugal
3 Secção de Micobactérias do Serviço de Patologia Clínica do Hospital de Pulido Valente, Lisboa, Portugal/Mycobacteria Section, Clinical Pathology Unit, Hospital de Pulido Valente, Lisboa, Portugal
4 Professora auxiliar convidada da Faculdade de Ciências Médicas da Universidade Nova de Lisboa. Assessora de Bacteriologia do Instituto Nacional de Saúde Dr. Ricardo Jorge. Departamento de Doenças Infecciosas, Instituto Nacional de Saúde Dr. Ricardo Jorge, Lisboa, Portugal/Assistant Guest Professor, School of Medical Sciences, Universidade Nova de Lisboa. Bacteriology assistant, Instituto Nacional de Saúde Dr. Ricardo Jorge. Department of Infectious Diseases, Instituto Nacional de Saúde Dr. Ricardo Jorge, Lisboa, Portugal
5 Professora auxiliar da Faculdade de Farmácia da Universidade de Lisboa. Investigadora visitante do Instituto Nacional de Saúde Dr. Ricardo Jorge, Centro de Patogénese Molecular, Faculdade de Farmácia da Universidade de Lisboa, Portugal. Departamento de Doenças Infecciosas, Instituto Nacional de Saúde Dr. Ricardo Jorge, Lisboa, Portugal/Guest Professor, School of Pharmacy, Universidade de Lisboa. Visiting Researcher, Instituto Nacional de Saúde Dr. Ricardo Jorge, Molecular Pathogenesis Centre, School of Pharmacy, Universidade de Lisboa, Portugal. Department of Infectious Diseases, Instituto Nacional de Saúde Dr. Ricardo Jorge, Lisboa, Portugal
Recebido para publicação/received for publication: 09.03.30
Aceite para publicação/accepted for publication: 09.04.17