Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Similares em
SciELO
Similares em
SciELO
Compartilhar
Revista de Ciências Agrárias
versão impressa ISSN 0871-018X
Rev. de Ciências Agrárias vol.41 no.3 Lisboa set. 2018
https://doi.org/10.19084/RCA17339
ARTIGO
Polyphasic characterization of forage legumes root nodule bacteria isolated from semiarid region in Brazil
Caracterização polifásica de bactérias de nódulos de leguminosas forrageiras isoladas da região semiárida do Brasil
Gérsika Fakirra de Oliveira Nunes1*, Kelly Alexsandra Souza Menezes1, Aline Araújo Sampaio1, Jakson Leite2, Paulo Ivan Fernandes-Júnior3, Sirando Lima Seido4, Jerri Édson Zilli5 and Lindete Míria Vieira Martins1*
1 Departamento de Tecnologia e Ciências Sociais, Universidade do Estado da Bahia, Juazeiro, BA, Brazil
2 Universidade Federal de Alagoas, Campus Arapiraca, Av. Manoel Severino Barbosa, Bom Sucesso, 57309-005, Arapiraca, Alagoas, Brazil
3 Embrapa Semiárido, BR 428, km 152, Petrolina, PE, Brazil, CEP: 56302-970, CP 23
4 Universidade Federal Rural de Pernambuco, Recife, PE, Brazil
5 Embrapa Agrobiologia, Rodovia BR-465, km 7. 23891-000, Seropédica, Rio de Janeiro, Brazil
(*E-mail: lindete.martins1@gmail.com)
ABSTRACT
Forage legumes are important resources in semiarid regions due to their abilities to adapt to soils with low fertility and fix nitrogen when associated with diazotrophic bacteria. Here, we applied a polyphasic approach to characterize a set of legume nodule isolates obtained from Clitoria ternatea and Stylosanthes capitata cultivated in the soils of a semiarid region of Brazil. A total of 126 bacterial isolates were obtained: 45 isolates from C. ternatea and 81 isolates from S. capitata. Nodulation tests revealed only ten isolates that nodulated their original host: six isolates from C. ternatea and four isolates from S. capitate. These ten legume nodule isolates were phenotypically and genotypically characterized. All isolates grew in fructose, glucose, sodium glutamate, maltose, xylose, and sucrose. Metabolic tests showed a relationship between tolerance to salt and high temperatures, where isolates that tolerated the highest salt concentration also tolerated the highest temperature. Three isolates showed amylolytic activity, and four isolates showed carboxymethyl cellulolytic activity. Streptomycin was the most limiting and nalidixic acid was the least limiting antibiotic to bacterial growth. Seven out of ten isolates were indol-acetic acid producers. Additionally, 16S rRNA gene partial sequencing enabled the identification of members of the genera Bacillus (1), Bradyrhizobium (4), Leifsonia (3), Microvirga (1), and Rhizobium (1). These data reveal phenotypically and genotypically diverse bacteria inhabiting the nodules of the forage legumes C. ternatea and S. capitata represent an important microbial source to protect new biotechnological products and improve forage legumes in semiarid regions.
Keywords: Biological nitrogen fixation, Bradyrhizobium, Clitoria ternatea, Stylosanthes capitata
RESUMO
Leguminosas forrageiras são importantes recursos na região semi-árida, devido a sua capacidade de adaptação a solos de baixa fertilidade e fixação de nitrogênio quando associados a bactérias diazotróficas. Aqui, aplicamos uma abordagem polifásica com o objetivo de caracterizar bactérias isoladas de nódulos obtidos de Clitoria ternatea e Stylosanthes capitata cultivadas em solos do semiárido brasileiro. O isolamento das bactérias produziu uma coleção de 126 isolados: 45 isolados de C. ternatea e 81 de S. capitata. Os testes de nodulação resultaram em apenas dez isolados que nodularam seu hospedeiro original: seis de C. ternatea e quatro de S. capitata. Esses dez isolados de nódulos de leguminosas tiveram suas características fenotípicas e genotípicos avaliadas. Todos os isolados cresceram em frutose, glicose, glutamato de sódio, maltose, xilose e sacarose. Testes metabólicos mostraram uma relação entre a tolerância ao sal e altas temperaturas; onde o isolado que tolerou a maior concentração de sal também tolerou a temperatura mais alta. Três isolados apresentaram atividade amilolítica e quatro foram capazes de produzir carboximetil celulolítico. A estreptomicina foi o antibiótico mais limitante para o crescimento bacteriano, e o ácido nalidíxico foi o menos limitante. Sete dos dez isolados eram produtores de IAA. O sequenciamento parcial do gene 16S rRNA permitiu identificar isolados como membros dos gêneros Bacillus (1), Bradyrhizobium (4), Leifsonia (3), Microvirga (1) e Rhizobium (1). Esses dados revelam diversidade fenotípicas e genotípicas de bactérias que habitam nódulos das forrageiras C. ternatea e S. capitata, e representam uma importante fonte microbiana para a prospecção de novos produtos biotecnológicos para promover melhor desenvolvimento das leguminosas forrageiras na região semiárida.
Palavras-chave: Fixação biológica de nitrogênio, Bradyrhizobium, Clitoria ternatea, Stylosanthes capitata
INTRODUCTION
The semiarid region of Brazil, with 24 million inhabitants (IBGE, 2017), is home to the largest population density among similar regions worldwide. The hot and dry climate, under an irregular rainfall regime concentrated in a few months of the year, imposes different risks to the agricultural activity and food security of farmers and their families.
Despite its great natural microbiological and genetic diversity, this ecosystem, which extends across eight states and occupies an area equivalent to 13% of the Brazilian territory, is the least studied to anticipate agricultural solutions to pressures of anthropogenic and temperature rise due to climate change.
Biological nitrogen fixation (BNF) has the potential for the development of technological resources in semiarid regions based on the ability of bacteria adapted to soil and climate to transform nitrogen into a product readily assimilable by plants. These diazotrophic bacteria contribute to the growth of most cultivable legume species (Moreira and Siqueira, 2006), and this ecological process is widely recognized for reducing the cost of production and the dependence of the farmer on industrialized inputs and is particularly important for tropical regions where the availability of this element in soils is often low and limiting to agricultural productivity.
The cultivation of forage legumes is an alternative to guarantee the supply of food during periods of scarcity. Clitoria ternatea L. and Stylosanthes capitata Vogel are legumes with great potential for cultivation in the semiarid regions of Brazil, due to their tolerance to drought, grazing, trampling, and water stresses (Lima et al., 2009; Mistura et al., 2010). In general, these legumes have been used in animal feeds and as components of green manures. Clitoria ternatea L. and Stylosanthes capitata Vogel also present good performance when symbiotically associated with nitrogen-fixing bacteria. However, no studies have reported the symbiotic bacteria associated with the nodules of these species in the soils of semiarid regions of Brazil.
Polyphasic studies that involve phenotypic and molecular characterization can increase the current knowledge on the physiology, taxonomy and potential selection of bacteria that show adaptations to the edaphoclimatic conditions peculiar to this territory, in addition to the biotechnological application of these microorganisms. Metabolic and genetic characterization has been one of the most used methods for initial classification and may play an important role in the identification of these bacteria.
Currently, no studies have yet been performed to evaluate the genetic and metabolic diversity of nitrogen-fixing bacteria associated with important legumes in semiarid regions (Leite et al., 2009; Menezes et al., 2016). Notably, knowledge of the diversity of this group of bacteria is limited but necessary considering the forage importance of these species. Therefore, the present study aims to identify the phenotypic and genotypic characteristics of the symbiotic nodules of Clitoria ternatea and Stylosanthes capitata bacteria grown on the semi-arid soils of Northeast Brazil.
MATERIAL AND METHODS
Bacterial isolation and nodulation tests
A survey of the root nodule bacteria associated with the forage legumes Clitoria ternatea L. and Stylosanthes capitata was performed by using a plant-trap experiment. To trap the bacteria, a greenhouse experiment was conducted by cultivating the two legume species in nine different soil samples. Each soil sample comprised ten subsamples collected from the surface horizon (0 - 20 cm) of a semiarid region of Brazil. After 60 days of cultivation, the plants were harvested, the nodules were collected, and subsequent bacterial isolation was performed in yeast mannitol agar (YMA) medium according to Vincent (1970).
Nodulation tests were performed under gnotobiotic conditions to confirm the capacity of the obtained isolates to nodulate the original host legume: 45 isolates from C. ternatea and 81 isolates from S. capitata. Plants of C. ternatea and S. capitata were cultivated in Leonard jars (Vincent, 1970) containing autoclaved (120ºC, 1 atm, 1 h) sand and vermiculite (1:1) substrate. The substrate was autoclaved twice on consecutive days. Five-day-old plants were inoculated with 1 ml (109 cells) of the tested isolate grown in yeast mannitol broth (Vincent, 1970). Each tested isolate was repeated thrice, and the jars were designed in a randomized block design in the greenhouse. Two uninoculated controls, without and with nitrogen application (75 ml plant-1 of NH4NO3), were added to assess contamination and plant growth. The plants were irrigated weekly with a nitrogen-free solution (Norris and Date, 1976) and distilled water when necessary. Due to the weak capacity of S. capitata to grow on a sterilized substrate, 7.5 mg of nitrogen was applied in the jars to promote the initial growth but not inhibit the nodulation of legumes (Guimarães et al., 2012). Nodulation (presence or absence, number and dry matter) and plant growth (shoot dry matter) data were recorded from 45-day-old plants.
The isolates that nodulate the original host were subjected to polyphasic characterization based on growth speed in YMA medium (fast: up to three days; and slow: over six days for colony formation), colony morphology (Table 1), growth on different carbon sources, enzyme activity (amylolytic and cellulolytic), production of indol-acetic acid, intrinsic antibiotic resistance, tolerance to different NaCl concentrations and temperature ranges, and genetic identification at the genus level by 16S rRNA gene partial sequencing.
Growth on different carbon sources
We used YMA medium (Vincent, 1970) to assess the capacity of the isolates to grow in the presence of different carbon sources (CS) by replacing mannitol with one of the following fourteen CS: arabinose, sodium acetate, citric acid, maleic acid, malic acid, succinic acid, casein, potassium citrate, fructose, glucose, sodium glutamate, maltose, xylose, and sucrose at 1% (w/v). The original YMA medium with mannitol was used as reference control. The isolates were incubated at 28ºC for up to ten days, and the growth was subsequently assessed. Positive capacity was attributed to isolates that grew in a manner similar to the reference control (mannitol).
Amylolytic and carboxymethyl cellulolytic activities
Amylolytic and cellulolytic activities were verified on YMA medium by replacing mannitol with maize starch (Maizena®) and carboxymethyl cellulose (CMC) at 1% (w/v), respectively. Bacterial growth was achieved after incubating three pure colonies in YM broth (Vincent, 1970) at 28ºC with shaking at 150 rpm for 5 days. Then, we inoculated 10 µL of bacterial culture broth at three equidistant points on 90 mm diameter-Petri dishes containing the tested medium (Fernandes Júnior et al., 2012). Amylolytic activity was detected by inoculating 5 mL of iodine dye solution (0.2% v/v) on seven-day-old incubated plates. Positive activity was considered when a smooth yellow halo formed around the colony, contrasting with the dark blue medium, according to Oliveira et al. (2006a).
Carboxymethyl cellulase activity was detected at 15 days after incubation by applying 5 mL of a Congo red solution (0.12% in 0.1 N KOH) and incubating for 5 minutes until the indicator was absorbed by the medium, and the excess was discarded. Then, 5 mL of a 10% acetic acid solution was applied, resulting in a change in medium color from blue to purple, enabling the appearance of a light halo around the colonies to indicate enzyme activity.
Enzyme activity was estimated as the ratio between the diameter of the halo and the diameter of the colony, expressed as an enzyme index (EI) (Hankin and Anagnostakis, 1975).
Indol-acetic acid (IAA) production
To determine the IAA produced by each isolate, the colorimetric method described by Sarwar and Kremer (1995) was employed with modifications. Twenty microliters of each isolate was inoculated onto 3 mL of yeast mannitol broth (YM broth) with 168 µg mL-1 of L-tryptophan and incubated at 28ºC under constant agitation of 120 rpm for five days. Subsequently, the optical density (OD) of each isolate was recorded at 540 nm, and adjusted to an OD540 of 0.5. Then, 2 ml aliquots of each bacterial isolate were centrifuged at 8000 rpm for 10 minutes, and 1200 µL of the supernatant was mixed with 800 µL of Salkowskis reagent (2% 0.5 FeCl3 in 35% HCLO4 solution) and incubated in the dark at room temperature. After 30 minutes, the optical density (OD) was recorded at 520 nm by using the Gold Spectrumlab 53 spectrophotometer. The concentrations of the indolic compounds produced by the isolates were estimated according to the standard curve equation generated with known concentrations of synthetic IAA.
Intrinsic antibiotic resistance
Intrinsic antibiotic resistance was analyzed by using discs impregnated with antibiotics (Bauer et al., 1966). The isolates were cultivated in YM broth (120 rpm for five days). Then, 100 μL of bacterial culture (109 cells) was spread onto Petri dishes containing YMA medium and a disc of the tested antibiotic. The antibiotics evaluated (Sensifarâ) were streptomycin (10 μg), rifampicin (5 μg), neomycin (30 μg), erythromycin (15 μg), vancomycin (30 μg), nalidixic acid (30 μg), gentamicin, ampicillin (10 μg) and chloramphenicol (30 μg). Each antibiotic was tested in triplicate on three different plates incubated at 28°C for seven days. The resistance or sensitivity was recorded based on the absence or presence of an inhibition zone of the tested antibiotic, respectively.
Growth in different NaCl concentrations and temperature ranges
Isolates were inoculated Petri dish containing YMA medium supplemented with different NaCl concentrations [1, 2 and 3% (w/v)]. The plates were incubated at 30 °C. For the high-temperature tolerance tests, the isolates were inoculated in YMA medium and incubated at 41, 43, 45, 47, 49, 51, 53, and 55 °C. In both assays, tolerance was recorded after seven days of incubation. Tolerance was attributed to isolates that grew along the streak, and non-tolerance was attributed to isolates that did not show visible growth.
Genetic characterization
Genomic DNA was extracted from cell cultures according to the Wizard® Genomic DNA extraction kit (#A1125, Promega). The 16S rRNA gene was amplified from the DNA through PCR in a final 50-µL volume containing: 1X buffer, MgCl2 1.5 μmol L-1, Taq DNA polymerase 1.75 U (Invitrogen cat. N.11615-010), dNTP 250 μmol L-1, and 0.2 μmol L-1 of each initiator. The primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-TACGGYTACCTTGTTACGACTT-3′) were used for the 16S rRNA amplification (Weisburg et al., 1991). PCR was performed on a PTC-200 Peltier Thermal Cycler (MJ Research, Waltham, EUA) with the following cycle program: initial denaturation at 95ºC for 3 minutes, followed by 30 cycles of denaturation at 95ºC for 1 minute, annealing at 55ºC for 1 minute, extension at 72ºC for 1 minute, and a final extension at 72ºC for 5 minutes. The amplification product was sent to Macrogen (South Korea) for purification and sequencing. The obtained sequences were compared by using the Blast tool (http://blast.ncbi.nlm.nih.gov) for the identification of correlated sequences and deposited at the National Center for Biotechnology Information GenBank database (http://www.ncbi.nlm.nih.gov/) as MH2O1287 – MH2O1296. Phylogenetic analysis was performed by using MEGA program version 4.0 (Tamura et al., 2007). Multiple sequence alignment was performed by using CLUSTALW (Thompson et al., 1994) implemented on the MEGA program with default parameters. The aligned sequences were used to select the best-fit substitution model measured by Bayesian information criteria (Schwarz, 1978). A phylogenetic tree was inferred by using the maximum likelihood method based on the Tamura 3-parameter model, as implemented in the bootstrap consensus tree inferred from 100 replicates (Felsenstein, 1985).
Experimental design and statistical analyses
All assays were conducted with a completely randomized triplicate design. The treatment means for the enzyme activity (amylolytic and carboxymethyl cellulolytic), and production of indoleacetic acid (IAA) were compared by the Scott-Knott test at 5% probability by using Sisvar 5.3 statistical analysis software (Ferreira, 2011).
RESULTS AND DISCUSSION
The bacterial isolation yielded a collection of 45 isolates from Clitoria ternatea and 81 isolates from Stylosanthes capitata nodules. The nodulation capacity was identified in only ten isolates: six isolates from C. ternatea and four isolates from S. capitata (Table 1). Then, these isolates were subjected to polyphasic characterization.
The legume nodule isolates showed different profiles regarding the use of carbon sources. All of the isolates were capable of growing in fructose, glucose, sodium glutamate, maltose, xylose, and sucrose. However, maleic acid, sodium acetate, and casein were the most limiting sources, each inhibiting the growth of four bacteria (Table 2).
There was a correlation between the growth time of the isolates and capability to metabolize C sources. Isolates with fast growth (263-2, 271-2, 273-2, 291-4 and 292-6) were capable of using a higher number of C sources; the growth of 263-2, 291-4, and 292-6 was limited only by maleic acid, whereas 271-2 and 273-2 grew in all sources evaluated. For isolates with slow growth, a higher selectivity in the use of C sources was observed, and the most limiting sources to these isolates were sodium acetate and casein.
According to Stowers (1985), fast-growing rhizobia are capable of using a wide range of sugars, salts, and organic acids, whereas slow-growing rhizobia are ineffective in using disaccharides, trisaccharides, and organic acids for growth. In addition, the use of sucrose was restricted to fast-growing rhizobia, and slow rhizobia were incapable of metabolizing this carbohydrate and other disaccharides, such as maltose. This finding was not confirmed in the present study, as sucrose and maltose were metabolized by all the isolates evaluated. However, organic salt sodium acetate limited the growth of the bacteria identified as slow-growing isolates and was used as a C source by all fast-growing isolates.
Previous studies have reported a high diversity in the use of carbohydrates by rhizobia isolates from different legumes (Küçük et al., 2006; Kumari et al., 2009; Razika et al., 2012). Rhizobium strains obtained from the nodules of Phaseolus vulgaris were capable of growing in fructose, galactose, glucose, mannitol, sucrose, starch, succinate, rhamnose, and malate, but were not able to grow in citrate and dulcitol (Küçük et al., 2006). In the present study, only two isolates did not use potassium citrate (263-3 and 391-9).
Kumari et al. (2009) evaluated the growth of five strains of Rhizobium isolates from nodules of the genus Indigofera on different carbon sources and observed maximum isolate growth for strains cultivated in media containing monosaccharides (glucose, galactose, arabinose, fructose, raffinose, and xylose), followed by mannitol, disaccharides (lactose, maltose, and sucrose), and polysaccharides (starch and cellulose).
The study of bacterial behavior, e.g., regarding the use of different carbon sources, is used to characterize and determine the diversity of bacteria. Each strain, cultivated in culture media containing different carbohydrates, shows better development with certain sugars than with others (Castellane and Lemos, 2007; Kumari et al., 2009). In addition, the metabolic diversity in the use of C might indicate the adaptability of microorganisms to different environmental conditions, influenced by the root exudates of plants (Shetta et al., 2011).
Regarding intrinsic antibiotic resistance, streptomycin (STR) was the most limiting antibiotic for bacterial growth, and all isolates were sensitive to this substance. However, nalidixic acid (NAL) did not limit the growth of any evaluated isolate (Table 2).
Antibiotic resistance analysis of five Rhizobium strains, three isolates from pigeonpea (Cajanus cajan) nodules and two isolates from cowpea (Vigna unguiculata) nodules, showed the high tolerance of bacteria to NAL and high sensitivity to STR (Fernandes & Fernandes et al., 2003). The authors also observed that their strains had high resistance to chloramphenicol (CLO), which was also observed by Keneni et al. (2010) when analyzing Rhizobium isolates of faba beans (Vicia faba L.) cultivated in soils from Ethiopia. In the present study, this antibiotic limited the growth of five isolates (263-2, 263-3, 291-4, 292-6, and 391-9).
Antibiotic resistance might be correlated with the production of mucus by Rhizobium isolates. In an analysis of three Rhizobium isolates, two isolates obtained from pigeonpea nodules and one isolate obtained from cowpea nodules, higher tolerance to antibiotics was observed in isolates that produced more mucus (Fernandes and Fernandes, 2000). This correlation was not observed in the present study: the most susceptible isolate produced a large quantity of mucus (263-2) and the most resistant isolate, 391-13, produced little mucus (Tables 1 and 2).
None of the isolates evaluated were capable of growing in culture medium supplemented with 3% NaCl (Table 2). Isolates 263-3, 391-9, 391-11, 391-12, and 391-13 were inhibited by 1% NaCl; isolates 291-4 and 292-6 grew in the medium supplemented with 1% NaCl but were inhibited by 2% NaCl. The isolates most tolerant to salinity were 263-2, 271-2, and 273-2, which grew in medium containing 2% NaCl (Table 2).
As for tolerance to high temperatures, there was variation among isolates (Table 2). Isolate 271-2 (Bacillus sp.) was able to grow at a temperature up to 53 °C, showing the highest thermal tolerance. However, the most sensitive isolates were 263-3 and 291-4, which did not grow at a temperature of 39°C.
Salinity and high temperatures are factors limiting the symbiotic process between rhizobia and legumes, affecting several important stages of the infection process, nodule formation, bacteroid differentiation, and nitrogen fixation. However, the adverse effect of these environmental conditions on the symbiotic process depends on the macro and micro-symbiont, and the selection of suitable partners is a mitigating factor for these environmental factors (Moreira and Siqueira, 2006).
Although in vitro responses to growth-limiting factors do not predetermine the bacterial behavior under field conditions, this type of analysis might be one of the first steps in selecting isolates tolerant to different environmental stresses, as these studies enable a larger number of evaluations in a shorter period of time with lower costs. Tolerance to different NaCl concentrations and maximum temperatures for legume-nodulating bacteria is well reported and diversified in the literature (Ali et al., 2009).
All isolates were capable of growing in the medium supplemented with starch; however, only three isolates showed extracellular amylolytic activity (271-2, 291-4 and 391-12). According to Oliveira et al. (2006a), the enzyme index (EI) is one of the most commonly used semiquantitative parameters to evaluate the ability of microorganisms to produce enzymes in solid media. The evaluation of enzyme-producing organisms directly correlates the diameter of the degradation halo with the degrading ability of microorganisms, and an EI ≥ 2.0 is recommended to consider a microorganism as a producer of enzymes in a solid medium.
None of the evaluated isolates increase the enzyme index (EI) to greater than 2 (Table 3), and the highest EI was obtained for isolate 291-4 (1.72). Previous studies observed EI values of 3.1 for cowpea rhizobial isolates (Oliveira et al., 2006b) and values of up to 3.5 for fast-growing isolates from pigeonpea (Cajanus cajan) (Fernandes Júnior et al., 2012). Kumari et al. (2010) reported the importance of the enzymes urease, protease, amylase, and gelatinase in the formation of nodules, stating that rhizobia that produce these enzymes are considered more effective in nodulation and N fixation.
All ten isolates evaluated grew in the medium supplemented with carboxymethyl cellulose (CMC) as the only carbon source. However, four isolates (291-4, 292-6, 391-9, and 391-11) were capable of extracellularly producing carboxymethyl cellulase (CMCase). Considering that an organism is a good producer when its EI is ≥ 2, isolate 292-6 stood out regarding this characteristic, with an EI equal to 2.70. Other isolates, 291-4 and 391-11, were intermediaries in the extracellular production of CMCase, and isolate 391-9 showed lower production of this enzyme (EI equal to 1.23) compared to that of the other isolates (Table 3).
The enzyme cellulase is essential to the symbiotic process between rhizobia and legumes, actively participating in the bacterial infection of root hairs and the subsequent release of bacteria to the infection thread inside host nodule cells. Thus, rhizobia capable of nodulating their host produce at least one type of cellulase at some point (Robledo et al., 2008).
Seven of the ten isolates tested produced IAA in medium supplemented with L-tryptophan. The IAA concentration ranged from 2.26 to 19.36 µM/mL; isolate 271-2, obtained from C. ternatea nodules, had the highest production, and isolate 391-13, derived from the nodules of Stylosanthes, had the lowest production. Isolates 291-4 and 391-9 produced the same amount of IAA, at 3.40 µM/mL (Table 3). Among the three isolates that did not produce IAA (263-3, 391-11, and 391-12), only one isolate was obtained from the nodules of C. ternatea (263-3).
In other studies (Chagas Júnior et al., 2009; Coatti et al., 2010; Sahasrabudhe, 2011), the production of IAA was associated with fast-growing isolates, mainly those belonging to the genus Rhizobium. These data corroborate the present findings, as the highest values of IAA were recorded in fast-growing isolates (Tables 1 and 3).
The ability to produce plant hormones is widely distributed among microorganisms in the soil, where approximately 80% of bacteria isolated from rhizospheres are IAA producers (Sahasrabudhe, 2011). The ability of rhizobia to process these plant hormones has been widely studied due to the suggestion that these organisms have a mechanism that provides a positive physiological response to their hosts (Machado et al., 2011). The performance of these in vitro assays for IAA production from L-tryptophan has shown good results in the selection of effective Rhizobium isolates (Stroschein, 2011).
BLAST analysis of the partial 16S rRNA gene sequences of the ten legume nodules isolates that show positive nodulation on the original host enabled the identification of Bacillus (271-2), Bradyrhizobium (263-3, 391-11, 391-12, and 391-13), Leifsonia (263-2, 291-4, and 292-6), Microvirga (391-9), and Rhizobium (273-2) genera represented in the collection (Table 1). Bradyrhizobium, Microvirga, and Rhizobium genera are well-known root legume nodule symbionts. Bacillus and Leifsonia isolates have been reported as nonrhizobial endophytes of the nodule microbiome (Muresu et al., 2008; Rajendran et al., 2008; Selvakumar et al., 2008, Cardoso et al., 2012). These two genera were isolated from C. ternatea nodules. This forage legume hosts nonrhizobial endophytes inside the root nodules (Aeron et al., 2015). The positive nodulation results of isolates 271-2 (Bacillus), and 263-3, 391-11, 391-12, and 391-13 (Leifsonia) must result from contamination or the mixture of culture with rhizobia under low density.
A phylogenetic analysis based on the 16S rRNA gene was performed with the isolates obtained in the present study, which belong to the known legume-nodulating bacteria genera Bradyrhizobium, Microvirga, and Rhizobium (Table 1). The maximum likelihood separated the isolates into five different lineages (Figure 1). S. capitata isolates 391-11, 391-12, and 391-13 were placed into the large subgroup I of the Bradyrhizobium genus (Menna et al., 2009), with 391-12 and 391-13 isolates correlated with Bradyrhizobium yuanmingense, and isolate 391-11 strongly affiliated (97% of bootstrap) with Bradyrhizobium kavangense. The S. capitata isolate 391-9 was phylogenetically close to Microvirga vignae BR3299T, a cowpea symbiont isolated from semiarid region in Brazil (Radl et al., 2014).
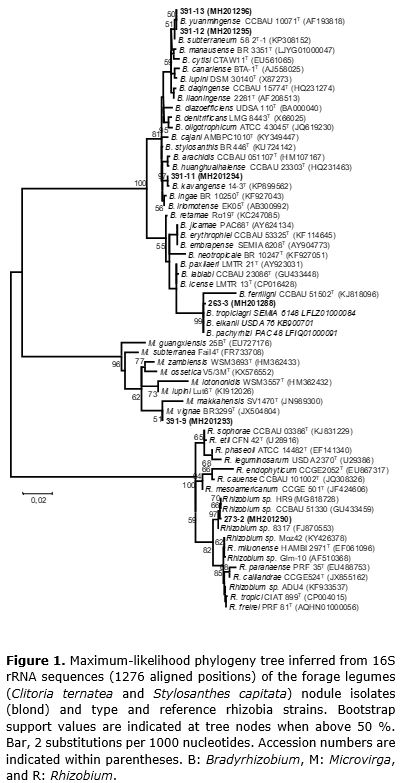
Stylosanthes spp. are typically nodulated by α-Proteobacteria (Date, 2010), mainly belonging to the genus Bradyrhizobium (Ramesh et al., 2004). Recently, β- Proteobacteria members were isolated from Stylosanthes nodules (Chaves et al., 2016), expanding the knowledge of the symbiotic capacity of Stylosanthes species. In the present study, we report the symbiotic capacity of S. capitata to form symbiosis with Microvirga, an α-Proteobacteria genus with members reported as legume-nodulating symbionts (Ardley et al., 2012; Radl et al., 2014; Msaddak et al., 2017a, b, Safronova et al., 2017).
In addition to its genotypic characteristic, isolate 391-9 showed phenotypic similarities with other representatives of the genus Microvirga, such as growth in YMA culture medium for a period longer than three days, inability to hydrolyze starch, susceptibility to the antibiotics streptomycin, rifampicin, erythromycin, and chloramphenicol, maximum temperature range tolerated (approximately 43°C), and carbon sources used (Tables 1, 2, and 3) (Ardley et al., 2012; Radl et al., 2014).
The C. ternatea isolate 263-3 fell into the cluster of the species B. tropiciagri, B. elkanii, and B. pachyrhizi (Figure 1), which compose subgroup II of the Bradyrhizobium genus (Menna et al., 2009). However, C. ternatea isolate 273-2 clustered within the genus Rhizobium with a group of strains isolated from tropical soils related to previously described species.
CONCLUSION
On the basis of these data, we conclude that Clitoria ternatea and Stylosanthes capitata nodules harbor symbiotic rhizobia and nonrhizobia endophytic bacteria that are phenotypically and genotypically diverse. Clitoria ternatea is nodulated by Rizobium and Bradyrizobium members and host nonrhizobia endophytes. Stylosanthes capitata root nodule symbionts primarily belong to Bradyrizobium within two lineages: B. yuanmingense and B. kavangense. Moreover, S. capitata is nodulated by Microvirga in the semiarid soils of Brazil.
REFERENCES
Aeron, A.; Chauhan, P.S.; Dubey, R.C.; Maheshwari, D.K.; Bajpai, V.K. (2015) - Root nodule bacteria from Clitoria ternatea L. are putative invasive nonrhizobial endophytes. Canadian Journal of Microbiology, vol. 61, n. 2, p. 131-142. https://doi.org/10.1139/cjm-2014-0483 [ Links ]
Ali, S.F.; Rawat, L.S.; Meghvansi, M.K. & Mahna, S.K. (2009) - Selection of stress-tolerant rhizobial isolates of wild legumes growing in dry regions of Rajasthan, India. ARPN Journal of Agricultural and Biological Science, vol. 4, n. 1, p. 13-18. [ Links ]
Ardley, J.K.; Parker, M.A.; De Meyer, S.E.; Trengove, R.D.; O'Hara, G.W.; Reeve, W.G.; Yates, R.J.; Dilworth, M.J.; Willems, A. & Howieson, J.G. (2012) – Microvirga lupini sp. nov., Microvirga lotononidis sp. nov. and Microvirga zambiensis sp. nov. are alphaproteobacterial root-nodule bacteria that specifically nodulate and fix nitrogen with geographically and taxonomically separate legume hosts. International Journal of Systematic and Evolutionary Microbiology, vol. 62, p. 2579-2588. http://dx.doi.org/10.1099/ijs.0.035097-0 [ Links ]
Bauer, A.W.; Kirby, W.M.M.; Sherris, J.C. & Turck, M. (1966) - Antibiotic susceptibility testing by it standardized disk method. American Journal of Clinical Pathology, vol. 45, n. 4, p. 493-496. [ Links ]
Cardoso, J.D.; Hungria, M. & Andrade, D.S. (2012) - Polyphasic approach for the characterization of rhizobial symbionts effective in fixing N2 with common bean (Phaseolus vulgaris L.). Applied Microbiology and Biotechnology, vol. 93, n. 5, p. 2035-2049. https://doi.org/10.1007/s00253-011-3708-2 [ Links ]
Castellane, T.C.L. & Lemos, E.G.M. (2007) - Composição de exopolissacarídeos produzidos por estirpes de rizóbios cultivados em diferentes fontes de carbono. Pesquisa Agropecuária Brasileira, vol. 42, n. 10, p. 1503-1506. http://dx.doi.org/10.1590/S0100-204X2007001000019 [ Links ]
Chagas Júnior, A.F.; Oliveira, L.A. & Oliveira, A.N. (2009) - Produção de ácido indolacético por rizóbios isolados de caupi. Revista Ceres, vol. 56, n. 6, p. 812-817. [ Links ]
Chaves, J. S.; Baraúna, A. C.; Mosqueira, C. A.; Gianluppi, V.; Zilli, J. E. & Silva, K. (2016) - Stylosanthes spp. from Amazon savanna harbour diverse and potentially effective rhizobia. Applied Soil Ecology, vol. 108, p. 54–61 [ Links ]
Coatti, G.C.; Andrade, D.S.; Cardoso, J.D. & Matos, M.A. (2010) - Produção de AIA e Diversidade Fenotípica de Estirpes Elite de Rizóbio Isoladas de Feijoeiro. Científica Ciências Biológicas e da Saúde, vol. 12, n. 1, p. 49-53. [ Links ]
Date, R.A. (2010) - Bradyrhizobium effectiveness responses in Stylosanthes hamata and S. seabrana. Tropical Grasslands, vol. 44, p. 141-157. [ Links ]
Felsenstein, J. (1985) - Confidence limits on phylogenies: An approach using the bootstrap. Evolution, vol. 39, n. 4, p. 783-791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x [ Links ]
Fernandes Júnior, P.I.; Lima, A.A.; Araújo, J.L.S.; Rumjanek, N.G. & Xavier, G.R. (2012) - Phenotypic diversity and amylolytic activity of fast growing rhizobia from pigeonpea [Cajanus cajan (L.) Millsp.]. Brazilian Journal of Microbiology, vol. 43 n. 4, p. 1604-1612. http://dx.doi.org/10.1590/S1517-83822012000400045 [ Links ]
Fernandes, M.F. & Fernandes, R.P.M. (2000) - Seleção inicial e caracterização parcial de rizóbios de tabuleiros costeiros quando associados ao guandu. Revista Brasileira de Ciência do Solo, vol. 24, n. 2, p. 321-327. http://dx.doi.org/10.1590/S0100-06832000000200009 [ Links ]
Fernandes, M.F.; Fernandes, R.P.M. & Hungria, M. (2003) - Seleção de rizóbios nativos para guandu, caupi e feijão-de-porco nos tabuleiros costeiros de Sergipe. Pesquisa Agropecuária Brasileira, vol. 38, n. 7, p. 835-842. http://dx.doi.org/10.1590/S0100-204X2003000700007 [ Links ]
Ferreira, D.F. (2011) - SISVAR: a computer statistical analysis system. Ciência e Agrotecnologia, vol. 35, n. 6, p. 1039–1042. http://dx.doi.org/10.1590/S1413-70542011000600001 [ Links ]
Guimarães, A. A.; Jaramillo, P. M. D.; Nóbrega, R. S. A.; Florentino, L. A.; Silva, K. B.; Moreira, F. M. S. (2012) - Genetic and Symbiotic Diversity of Nitrogen-Fixing Bacteria Isolated from Agricultural Soils in the Western Amazon by Using Cowpea as the Trap Plant. Applied and Environmental Microbiology, vol. 78, p.6726-6733. [ Links ]
Hankin, L. & Anagnostakis, S.L. (1975) - The use of solid media for detection of enzymes production by fungi. Mycologia, vol. 67, n. 3, p. 597-607. http://dx.doi.org/10.2307/3758395 [ Links ]
IBGE (2017) - Estimativa da população dos municípios do semiárido brasileiro. Instituto Brasileiro de Geografia e Estatística. [cit. 2018.03.26]. <https://sidra.ibge.gov.br> [ Links ]
Keneni, A.; Assefa, F. & Prabu, P.C. (2010) - Characterization of acid and salt tolerant rhizobial strains isolated from faba bean fields of Wollo, Northern Ethiopia. Journal of Agricultural Science and Technology, vol. 12, n. 3, p. 365-376. [ Links ]
Küçük, Ç.; Kivanç, E. & Kinaci, E. (2006) - Characterization of Rhizobium sp. isolated from bean. Turkish Journal of Biology, vol. 30, p. 127-132. [ Links ]
Kumari, B.S.; Ram, M. R. & Mallaiah, K.V. (2009) - Studies on exopolysaccharide and indole acetic acid production by Rhizobium strains from Indigofera. African Journal of Microbiology Research, vol. 3, n. 1, p. 10-14. [ Links ]
Kumari, B.S.; Ram, M.R. & Mallaiah, K.V. (2010) - Studies on nodulation, biochemical analysis and protein profiles of Rhizobium isolated from Indigofera species. Malaysian Journal of Microbiology, vol. 6, n. 2, p. 133-139. http://dx.doi.org/10.21161/mjm.20109 [ Links ]
Leite, J.; Seido, S.L.; Passos, S.R.; Xavier, G.R.; Runjaneck, N.G. & Martins, L.M.V. (2009) - Biodiversity of rhizobia associated with cowpea cultivars in soils of the lower half of the São Francisco River Valley. Revista Brasileira de Ciência do Solo, vol. 33, n. 5, 1215-1226. http://dx.doi.org/10.1590/S0100-06832009000500015 [ Links ]
Lima, G.F.C.; Araújo, G.G.L. & Maciel, F.C. (2009) - Produção e conservação de forragens para sustentabilidade dos rebanhos 1 caprinos e ovinos na base da agricultura familiar. Revista Tecnologia & Ciência Agropecuária, vol. 3, n. 4, p. 43-53. [ Links ]
Machado, R.G.; Sá, E.L.S.; Damasceno, R.G.; Hahn, L.; Almeida, D.; Moraes, T.; Camargo, F.A.O. & Reartes, D.S. (2011) - Promoção de crescimento de Lotus corniculatus L. e Avena strigosa Schreb pela inoculação conjunta de Trichoderma harzianum e rizóbio. Ciência e Natura UFMS, vol. 33, n. 2, p. 111-126. http://dx.doi.org/10.5902/2179460X9365 [ Links ]
Menezes, K.A.S.; Nunes, G.F.O.; Sampaio, A.A.; Silva, A.F.; Souza, L.S.B.; Gava, C.A.T.; Martins, L.M.V. & Fernandes-Junior, P.I. (2016) - Diversity of new root nodule bacteria from Erythrina velutina Willd., a native legume from the dry forest Caatinga (Northeastern, Brazil). Revista de Ciências Agrárias, vol. 39, n. 2, p. 222-33. http://dx.doi.org/10.19084/RCA15050 [ Links ]
Menna, P.; Barcellos, F. G.; Hungria, M. (2009) - Phylogeny and taxonomy of a diverse collection of Bradyrhizobium strains based on multilocus sequence analysis of the 16S rRNA gene, ITS region and glnII, recA, atpD and dnaK genes. International Journal of Systematic and Evolutionary Microbiology, vol. 59, n. 12, p. 2934-2950. http://dx.doi.org/10.1099/ijs.0.009779-0 [ Links ]
Mistura, C.; Oliveira, J.M.; Souza, T.C.; Vieira, P.A.S.; Lima, A.R.S.; Oliveira, F.A.; Dourado, D.L. & Silva, R.M. (2010) – Adubação orgânica no cultivo da Cunhã na região semiárida do Brasil. Revista Brasileira de Saúde e Produção Animal, vol. 11, n. 3, p. 581-594. [ Links ]
Moreira, F.M.S. & Siqueira, J.O. (2006) - Microbiologia e Bioquímica do Solo. 2ª ed. UFLA, Lavras, 729 p. [ Links ]
Msaddak, A.; Durán, D.; Rejili, M.; Mars, M.; Ruiz-Argüeso, T.; Imperial J.; Palacios, J.M. & Rey, L. (2017a) - Diverse Bacteria Affiliated with the Genera Microvirga, Phyllobacterium, and Bradyrhizobium Nodulate Lupinus micranthus Growing in Soils of Northern Tunisia. Applied and Environmental Microbiology, v. 83, n. 6, art. e02820-16. https://doi.org/10.1128/AEM.02820-16 [ Links ]
Msaddak, A.; Rejili, M.; Durán, D.; Rey, L.; Imperial J.; Palacios, J.M.; Ruiz-Argüeso, T. & Mars, M. (2017b) - Members of Microvirga and Bradyrhizobium genera are native endosymbiotic bacteria nodulating Lupinus luteus in Northern Tunisian soils. FEMS Microbiology Ecology, vol. 93, n. 6, fix068. https://doi.org/10.1093/femsec/fix068 [ Links ]
Muresu, R.; Polone, E.; Sulas, L.; Baldan, B.; Tondello, A.; Delogu, G.; Cappuccinelli, P.; Alberghini, S.; Benhizia, Y.; Benhizia, H.; Benguedouar, A.; Mori, B.; Calamassi, R.; Dazzo, F.B. & Squartini, A. (2008) - Coexistence of predominantly nonculturable rhizobia with diverse, endophytic bacterial taxa within nodules of wild legumes. FEMS Microbiology Ecology, vol. 63, n. 3, p. 383-400. http://dx.doi.org/10.1111/j.1574-6941.2007.00424.x [ Links ]
Norris, D. O.; Date, R.A. (1976) - Legume Bacteriology. In: SHAM, N. H.; BRYAN, W. W. (ed). Tropical Pasture Research – Principles and Methods. Hurley: COMMONWEALT BUREAU OF PASTURES AND FIELD CROPS. p. 134-174. (bulletin, 51). [ Links ]
Oliveira, A.N.; Oliveira, L.A.; Andrade, J.S. & Chagas Júnior, A.F. (2006a) - Atividade enzimática de isolados de rizóbia nativos da amazônia central crescendo em diferentes níveis de acidez. Ciência e Tecnologia de Alimentos, vol. 26, n. 1, p. 204-210. http://dx.doi.org/10.1590/S0101-20612006000100032 [ Links ]
Oliveira, A.N.; Oliveira, L.A.; Andrade, J.S. & Chagas Júnior, A.F. (2006b) - Enzimas hidrolíticas extracelulares de isolados de rizóbia nativos da Amazônia central, Amazonas, Brasil. Ciência e Tecnologia de Alimentos, vol. 26, n. 4, p. 853-860. http://dx.doi.org/10.1590/S0101-20612006000400022 [ Links ]
Radl, v.; Simões-Araújo, j.l.; Leite, j.; Passos, s.r.; Martins, l.m.v.; Xavier, g.r.; Rumjanek, n.g.; Baldan, J.i. & Zilli, J.E. (2014) - Microvirga vignae sp. nov., a root nodule symbiotic bacterium isolated from cowpea grown in semi-arid Brazil. International Journal of Systematic and Evolutionary Microbiology, vol. 64, n. 3, p.725–730. http://dx.doi.org/10.1099/ijs.0.053082-0 [ Links ]
Rajendran, G.; Sing, F.; Desai, A.J. & Archana, G. (2008) - Enhanced growth and nodulation of pigeon pea by co-inoculation of Bacillus strains with Rhizobium spp. Bioresource Technology, vol. 99, n. 11, p. 4544-4550. http://dx.doi.org/10.1016/j.biortech.2007.06.057 [ Links ]
Ramesh, C.R.; Kulkarni, J.H. & Desale, J.S. (2004) - Response of Stylosanthes seabrana to Bradyrhizobium inoculation in India. In: Chakraborty, S. (Ed.) - High-yielding anthracnose-resistant Stylosanthes for agricultural systems. Camberra: Aciar, Cap. 14, p. 159-162. [ Links ]
Razika, G.; Amira, B.; Yacine, B. & Ammar, B. (2012) - Influence of carbon source on the production of exopolysaccharides by Rhizobium sullae and on the nodulation of Hedysarum coronarium L. legume. African Journal of Microbiology Research, vol. 6, n. 30, p. 5940-5946. http://dx.doi.org/10.5897/AJMR12.393 [ Links ]
Robledo, M.; Jiménez-Zurdo, J.I.; Velázquez, E.; Trujillo, M.E.; Zurdo-Piñeiro, J.L.; Ramírez-Bahena, M.H.; Ramos, B.; Díaz-Mínguez, J.M.; Dazzo, F.; Martínez-Molina, E. & Mateos, P.F. (2008) - Rhizobium cellulase CelC2 is essential for primary symbiotic infection of legume host roots. Proceedings of the National Academy of Sciences of the United States of America, vol. 105, p. 7064-7069. http://dx.doi.org/10.1073/pnas.0802547105 [ Links ]
Safronova, V.I.; Kuznetsova, I.G.; Sazanova, A.L.; Belimov, A.A.; Andronov, E.E.; Chirak, E.R.; Osledkin, Y.S.; Onishchuk, O.P.; Kurchak, O.N.; Shaposhnikov, A.I.; Willems, A. & Tikhonovich, I.A. - (2017) - Microvirga ossetica sp. nov., a species of rhizobia isolated from root nodules of the legume species Vicia alpestris Steven. International Journal of Systematic and Evolutionary Microbiology, vol. 67, p. 94-100. http://dx.doi.org/10.1099/ijsem.0.001577 [ Links ]
Sahasrabudhe, M. (2011) - Screening of rhizobia for indole acetic acid production. Annals of Biological Research, vol. 2, n. 4, p. 460-468. [ Links ]
Sarwar, M. & Kremer, R.J. (1995) - Determination of bacterially derived auxins using a microplate method. Letters in Applied Microbiology, vol. 20, n. 5, p. 282-285. http://dx.doi.org/10.1111/j.1472-765X.1995.tb00446.x [ Links ]
Schwarz, G. (1978) - Estimating the dimension of a model. The Annals of Statistics, vol. 6, n. 2, p. 461–464. [ Links ]
Selvakumar, G.; Kundu, S.; Gupta, A.D.; Shouche, Y.S. & Gupta, H.S. (2008) - Isolation and characterization of nonrhizobial plant growth promoting bacteria from nodules of kudzu (Pueraria thunbergiana) and their effect on wheat seedling growth. Current Microbiology, vol. 56, n. 2, p. 134–139. http://dx.doi.org/10.1007/s00284-007-9062-z [ Links ]
Shetta, N.D.; Al-Shaharani, T.S. & Abdel-Aal, M. (2011) - Identification and Characterization of Rhizobium Associated with Woody Legume Trees Grown under Saudi Arabia Condition. American-Eurasian Journal of Agricultural & Environmental Sciences, vol. 10, n. 3, p. 410-418. [ Links ]
Stowers, M.D. (1985) - Carbon metabolism in Rhizobium species. Annual Review of Microbiology, vol. 39, p. 89-108. http://dx.doi.org/10.1146/annurev.mi.39.100185.000513 [ Links ]
Stroschein, M.R.D. (2011) - Seleção de rizóbios e efeito do nitrogênio na simbiose com alfafa e cornichão. Tese de doutoramento. Porto Alegre, Universidade Federal do Rio Grande do Sul. 140 p. [ Links ]
Tamura, K.; Dudley, J., Nei, M. & Kumar, S. (2007) - MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution, vol. 24, n. 8, p. 1596-1599. http://dx.doi.org/10.1093/molbev/msm092 [ Links ]
Thompson, J.D.; Higgins, D.G. & Gibson, T.J. (1994) - CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research, vol. 11, n. 22, p. 4673-4680. [ Links ]
Vincent, J.M. (1970) - A manual for the practical study of root nodule bacteria. IBP Handbook, n. 15, Blackwell, Oxford, 164 p. [ Links ]
Weisburg, W.G.; Barns, S.M.; Pelletier, D.A. & Lane, D.J. (1991) - 16S ribosomal DNA amplification for phylogenetic study. Journal of Bacteriology, vol. 173, n. 2, p. 697‑703. https://doi.org/10.1128/jb.173.2.697-703.1991 [ Links ]
Received/recebido: 2017.12.26
Received in revised form/recebido em versão revista: 2018.04.24
Accepted/aceite: 2018.05.02