Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Similares em
SciELO
Similares em
SciELO
Compartilhar
Revista de Ciências Agrárias
versão impressa ISSN 0871-018X
Rev. de Ciências Agrárias vol.41 no.spe Lisboa dez. 2018
https://doi.org/10.19084/RCA.17067
ARTIGO
Accessing the link between subtilases and lipid signalling events in grapevine downy mildew resistance
Elucidando a ligação entre subtilases e a sinalização lipídica na resistência da videira ao míldio
Joana Figueiredo1,2,3, Gonçalo Laureano1, Clemente da Silva1, Marisa Maia1,2,3, Marta Sousa Silva2,3, e Andreia Figueiredo1,*
1 Biosystems & Integrative Sciences Institute (BioISI), Faculdade de Ciências, Universidade de Lisboa, Lisboa, Portugal
2 Laboratório de FTICR e Espectrometria de Massa Estrutural, Faculdade de Ciências, Universidade de Lisboa, Lisboa Portugal
3 Centro de Química e Bioquímica, Faculdade de Ciências, Universidade de Lisboa, Lisboa, Portugal
(*E-mail: aafigueiredo@fc.ul.pt)
ABSTRACT
Subtilases are serine peptidases involved in several plant biological functions, however one of their most important participation is in the response to biotic and abiotic stresses. Subtilases have been linked to hormone-associated signalling, like the jasmonic acid (JA) pathway, that is particularly related with defence responses against necrotrophic fungus and herbivores. In grapevine, recent studies have implicated the JA pathway in response to Plasmopara viticola, a biotrophic oomycete. Our more recent results showed an increased expression of grapevine subtilases after P. viticola infection and JA elicitation at first hours after plant stress induction. Our aim is to deeply uncover subtilase participation in hormone-signalling pathway associated to grapevine-P. viticola interaction.
Keywords: Subtilases, Jasmonic acid, Vitis vinifera, Plasmopara viticola
RESUMO
Subtilases são peptidases serínicas envolvidas em várias funções biológicas das plantas, sendo uma das mais importantes funções a sua participação na resposta a stresses bióticos e abióticos. As subtilases têm sido associadas à sinalização mediada por hormonas, como a via do ácido jasmónico, que está particularmente associada à resposta de defesa contra fungos necrotróficos e herbívoros. Na videira, estudos recentes têm relacionado esta via na resposta da videira ao oomicete obrigatório Plasmopara viticola. Os nossos resultados mais recentes mostraram uma expressão aumentada das subtilases na videira após infeção com o míldio e elicitação com o ácido jasmónico nas primeiras horas após a indução do stress na planta. O nosso objetivo é elucidar a participação das subtilases na via de sinalização hormonal associada à interação da videira com o míldio.
Palavras-chave: Subtilases, Ácido Jasmónico, Videira, Plasmopara viticola
INTRODUCTION
Subtilisin-like proteases, commonly known as subtilases, are the second largest family of serine peptidases present in all kingdoms and with a wide range of biological functions. In plants, subtilases participate not only in normal protein turnover and plant development (e.g. seeds and fruits’ development), cell wall modification, and processing of peptide growth factors, but also in plant defence response against abiotic and biotic stresses (reviewed in Figueiredo et al., 2018). The involvement of subtilases in plant defence response became one of the most discussed and important topics in plant-pathogen interactions during the last decade. Only in the past five years, the scientific community witnessed an exponential increase of research works focused on subtilases and their role in plant defence against the most diverse pathogens or environmental stresses (Duan et al., 2016; Fan et al., 2016; Ekchaweng et al., 2017).
Subtilases are characterized by a conserved peptidase S8 domain that comprise a catalytic triad with an aspartate (Asp), a histidine (His) and a serine (Ser) amino acid residues (Dodson and Wlodawer, 1998). Within this catalytic domain, some subtilases have a protease-associated domain (PA) implicated in protein-protein interaction and substrate recognition. PA is also responsible for the homodimerization of the protein, to activate it when necessary (Siezen and Leunissen, 1997). Another conserved domain within plant subtilases is the inhibitor domain I9 or pro-domain, responsible for the enzyme inactivation preventing the access of the substrate to the active site, until the activation of the subtilase (Zhu et al., 1989; Li and Inouye, 1994). Some subtilases also contain a fibronectin (Fn) III-like domain, required for its activity, but it is dispensable in others (Rawlings and Salvesen, 2013). Most plant subtilases are directed to the secretory pathway and present a signal peptide that is cleaved as a prerequisite for enzyme maturation. Another important characteristic of the plant subtilases is glycosylation, a post-translational modification that regulates their activity. The most important protein glycosylation form is N-linked, formed by the covalent attachment of asparagine (Asn)-linked carbohydrates to the protein. These features are highly conserved within plants and most subtilases need them to reach their action site and become activated to perform their function properly (Figueiredo et al., 2018).
Despite several published studies around structure and biological functions of plant subtilases, little is known about their substrates, interaction partners and action mechanism in plant-pathogen interactions. Four decades after the discovery of the first subtilase by Lindstrom-Lang and Ottesen (1947), the first plant subtilase’ substrate, systemin, was identified in tomato (Schaller and Ryan, 1994). Systemin is a travelling peptide hormone with 18 amino acid residues, derived from proteolytic processing of a 200-amino-acid precursor protein named prosystemin. This peptide hormone is biologically active in low doses and participates in the signalling processes associated with the initiation of the systemic wound-induced defence response (Beloshistov et al., 2018). Also in tomato, the leucine-rich repeat (LRR) protein was described as another subtilase’ substrate and it was suggested its involvement in the mediation of molecular recognition and/or protein interaction processes to initiate immune signalling (reviewed in Schaller et al., 2018). A link between these two subtilase’ substrates was identified and it is currently hypothesized that, after prosystemin processing, the delivered systemin is recognized at the cell surface by a LRR receptor-like kinase which induces, at the site of wounding, the jasmonic acid (JA) synthesis pathway as a prerequisite for systemic defence gene induction (reviewed in Sun et al., 2011). Also, very recently, it was found that the prosystemin is processed into systemin by a specific type of subtilase named phytaspase (Beloshistov et al., 2018). Phytaspases are a group of subtilases with aspartate cleavage specificity that have an aspartate residue at the pro-domain–peptidase S8 domain junction, a feature that serves as a phytaspase signature within the plant subtilase family. Phytaspases have been associated with programmed cell death (PCD) in plants exposed to biotic and abiotic stresses (Chichkova et al., 2017). In tomato, under normal conditions, the phytaspase is located at the apoplast compartment, however, upon PCD-inducing stress such as pathogen attack, phytaspase is translocated to the cell interior where it cleaves the prosystemin delivering the systemin peptide hormone (Chichkova et al., 2010). Systemin will migrate to the wounding site, interact with the LRR receptor-like kinase and act as a paracrine signal inducing the octadecanoid pathway activation for the jasmonic acid biosynthesis. JA will work as an endocrine signal propagating the wound response through the activation of defence-related genes and production of protease inhibitors, protecting the plant from further attack (Beloshistov et al., 2018). In Arabidopsis thaliana, LRR protein was also identified as one of the SBT3.3 subtilase’ substrates (Tornero et al., 1996). Ramírez and co-workers suggested the involvement of the A. thaliana SBT3.3 on the LRR-containing proteins’ cleavage, including pattern recognition receptors (PRR) as PRR-type receptors and activation of plasma membrane receptors and consequent downstream signalling processes (Ramírez et al., 2013).
Grapevine (Vitis vinifera L.) is a fruit plant cultivated worldwide that presents a huge economic importance in the wine industry, particularly in Portugal, where it accounted for 680 million euro of exports in 2016 (OIV, 2017). One of the diseases that affects grapevine is the downy mildew, caused by the biotroph oomycete Plasmopara viticola (Berk. et Curt.) and De Toni, and causing high losses at each crop season (Buonassisi et al., 2017). The grapevine-P. viticola pathosystem has been widely studied (reviewed in Buonassisi et al., 2017) and several evidences were presented regarding both subtilase and JA involvement in the establishment of an incompatible interaction (Figueiredo et al., 2016; Guerreiro et al., 2016). It is widely accepted that JA signalling pathway plays a central role in plant defence against necrotrophic pathogen and insects, through the activation of the defence-related genes expression culminating in the accumulation of secondary metabolites and pathogenesis-related proteins (Glazebrook, 2005). However, only very recently it was also associated to plant resistance against biotrophs, namely in the grapevine-Plasmopara viticola interaction (Figueiredo et al., 2016; Guerreiro et al., 2016).
Regarding the involvement of subtilases in this pathosystem, our previous results suggest that some members of this serine protease family may be involved in the establishment of an effective defence response leading to the establishment of the incompatible interaction between grapevine and P. viticola (Figueiredo et al., 2016).We observed that these subtilases are constitutively expressed in resistant genotypes and highly induced after P. viticola inoculation, especially in the first hours after infection (Figueiredo et al., 2016). One of the subtilases (VvSBT4.19 isoform X1) showed a high expression increase at 6 hours after inoculation with this pathogen. These two studies raised the hypothesis of a possible involvement of subtilases with JA signalling pathway, considering that the first described subtilase’ substrates are associated with the activation of this pathway. Indeed, at the first hours of establishment of the P. viticola infection both gene expression of some subtilases and JA signalling pathway are activated. However, the mechanism by which these two features are linked in this pathosystem remains to be unveiled. In cotton plants, studies have described a subtilase (GbSBT1) that is activated and the protein moved to the cytoplasm after plant treatment with JA and ethylene, suggesting that JA signalling is required for plant resistance against Verticillium dahliae and suggesting a possible involvement of subtilases in this process (Duan et al., 2016). In Sorghum bicolor elicited with methyl jasmonate (MeJA) it was also observed an increase of expression of a subtilase gene (Salzman et al., 2005).
Based on our previous results of gene expression of grapevine subtilases upon P. viticola inoculation, we have selected the two more expressed genes (VvSBT4.19 isoform X1 and VviSBT5.3a) at the first hours post inoculation (6 and 24 hpi) and we have accessed its expression in the same grapevine genotypes upon JA elicitation. The results obtain in these two conditions, P. viticola inoculation and JA elicitation, are where discussed.
MATERIAL AND METHODS
Plant Material
Two Vitis vinifera genotypes, Regent’ and Trincadeira’ (tolerant and susceptible to Plasmopara viticola, respectively) were selected to assess subtilase gene expression after elicitation with jasmonic acid . Wood cuttings from the two genotypes were obtained at INIAV- Estação Vitivinícola Nacional (Dois Portos, Portugal) and grown in 2.5 L pots in universal substrate under controlled conditions in a climate chamber at natural day/night rhythm, relative humidity 60% and a photosynthetic photon flux density of 300 µmol m-2 s-1.
Elicitation Experiments
Grapevine leaves were elicited with 1mM JA (Sigma Aldrich) in 0.05% (v/v) TWEEN 20 solution, by spraying the entire plant. Control plants were sprayed with a 0.05% (v/v) TWEEN 20. The second and third fully expanded leaves beneath the shoot apex were harvested at 6 and 24 hours post elicitation (hpe), immediately frozen in liquid nitrogen and stored at −80°C. Three biological replicates were collected, being each biological replicate a pool of three leaves from three different plants.
Quantitative Real-Time PCR
Total RNA was isolated from frozen leaves with the Spectrum Plant Total RNA Kit (Sigma-Aldrich, USA), according to manufacturer's instructions. Residual genomic DNA was digested with DNase I (On-Column DNase I Digestion Set, Sigma-Aldrich, USA). RNA purity and concentration were measured at 260/280 nm using a spectrophotometer (NanoDrop-1000, Thermo Scientific) while RNA integrity was verified by agarose gel electrophoresis (1.2% agarose in TBE buffer). Genomic DNA (gDNA) contamination was checked by qPCR analysis of a target on the crude RNA (Vandesompele et al., 2002). Complementary DNA (cDNA) was synthesized from 2.5 µg of total RNA using RevertAid®H Minus Reverse Transcriptase (Fermentas, Ontario, Canada) anchored with Oligo(dT)23 primer (Fermentas, Ontario, Canada), according to manufacturer's instructions.
Quantitative real-time PCR experiments were carried out using Maxima SYBR Green qPCR Master Mix (2×) kit (Fermentas, Ontario, Canada) in a StepOne Real-Time PCR system (Applied Biosystems, Sourceforge, USA). A final concentration of 2.5 mM MgCl2 and 0.2 μM of each primer were used in 25 μL volume reactions, together with 4 μL of cDNA as template. Primer sequences and reaction details are provided in Table 1. Thermal cycling for all genes started with a denaturation step at 95°C for 10 minutes followed by 40 cycles of denaturation at 95°C for 15 seconds and annealing at the appropriate temperature for 30 seconds. Each set of reactions included a control without cDNA template. Dissociation curves were used to analyse non-specific PCR products. Three biological replicates and two technical replicates were used for each sample. Gene expression (fold change) was calculated as described in Hellemans et al. (2007). The reference genes used for the normalization were the previously described in Monteiro et al. (2013). Statistical significance (p < 0.05) of gene expression was determined by the Mann–Whitney U test using IBM® SPSS® Statistics version 23.0 software (SPSS Inc., USA).
RESULTS AND DISCUSSION
In 2016, the subtilase gene family was characterized in grapevine and the gene expression of several subtilases, predictably associated to plant defence, was accessed in grapevine-Plasmopara viticola pathosystem (Figueiredo et al., 2016). In these results, two subtilases were highlighted due to increased expression after 6 hours of infection with the P. viticola. The most interesting was the VviSBT4.19 isoform X1 subtilase gene (XM_010660203.1) that presented a 300-fold increase of gene expression at 6 hpi in the resistant genotype (R6hpi), (Figueiredo et al., 2016; Figure 1A). The second subtilase, VviSBT5.3a (XM_002266692.3), showed an increase of gene expression much less pronounced at this time-point in the same genotype (Figueiredo et al., 2016; Figure 1A). Despite remaining up-regulated at 24 hpi, both genes’ expression decreases when comparing to 6 hpi. In the resistant genotype, the response to the P. viticola inoculation is associated to the expression regulation of these two subtilases. In the susceptible genotype, the most significant gene expression increase was from VviSBT4.19 isoform X1 subtilase gene at 24 hpi (Figure 1A).
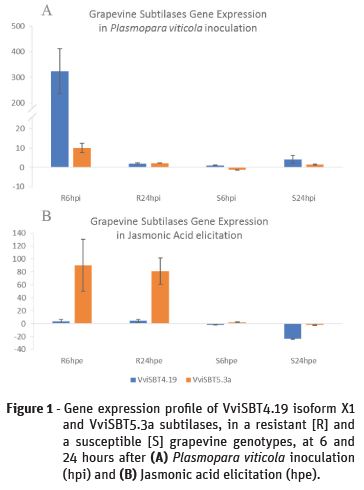
When comparing both incompatible and compatible interactions, our results suggest that the expression induction of both subtilase genes analysed presents a delay in the susceptible genotype. A faster activation of these subtilases in the resistant genotype may be related to the successful establishment of the defence strategy against the invading pathogen.
Based on the previous results, the gene expression of the VviSBT4.19 isoform X1 and VviSBT5.3a grapevine subtilases was analysed after grapevine elicitation with JA to access its response after increasing the grapevine defences. The results, contrarily to the P. viticola inoculation, showed that the subtilase gene with the higher up-regulation was the VviSBT5.3a, showing a 90 and 80-fold gene expression increase at 6 and 24 hpe, respectively, in the resistant genotype (Figure 1B). In the susceptible genotype we may highlight the accentuated down-regulation of the VviSBT4.19 isoform X1 subtilase gene at 24 hpe (Figure 1B). These preliminary results suggest that when the plant's defences are activated by elicitation with JA, the gene expression of specific subtilases, such as VviSBT5.3a, is extremely enhanced and enduring. This lead us to hypothesize that depending on the stimulus, the response of grapevine subtilases will be different. Our preliminary results present good clues to subtilases and JA-associated signalling activation of a defence response in grapevine after P. viticola inoculation.
CONCLUSIONS
The role of subtilases in plant defence response against the most diverse biotic stimulus have been extremely discussed in the last years. More recently an effort has been made to understand if there is any connection between these serine proteases and hormone-associated signalling, which plays a key role in plant defence. In grapevine, this connection is being made now and our first results point out for a possible link between subtilases and JA signalling pathway. When grapevines are elicited with this phytohormone, the gene expression of some subtilases increases. More studies must be conducted to confirm our hypotheses and fully understand the relation between subtilases and JA signalling pathway and the subtilases’ role in grapevine defence response against P. viticola attack.
Currently, the most used prevention approach to control grapevine fungal diseases, like downy mildew, is the extensive pesticide application each growing season, not always effective, prejudicial to human health and with consequent impact in the economy and environment. The fully comprehension of this plant-pathogen interaction is crucial for the discovery of grapevine host molecules that may be used to prevent pathogen attack or decrease its impact, helping to improve the quality of vineyards and wine, taking in mind a healthier and sustainable agriculture.
References
Beloshistov, R.E.; Dreizler, K.; Galiullina, R.A.; Tuzhikov, A.I.; Serebryakova, M.V.; Reichardt, S.; Shaw, J;, Taliansky, M.E.; Pfannstiel, J.; Chichkova, N.V.; Stintzi, A.; Schaller, A. and Vartapetian, A. (2018) - Phytaspase-mediated precursor processing and maturation of the wound hormone systemin. New Phytologist, vol. 218, n. 3, p. 1167–1178. https://doi.org/10.1111/nph.14568 [ Links ]
Buonassisi, D.; Colombo, M.; Migliaro, D.; Dolzani, C.; Peressotti, E.; Mizzotti, C.; Velasco, R.; Masiero, S.; Perazzolli, M. and Vezzulli, S. (2017) - Breeding for grapevine downy mildew resistance: a review of “omics” approaches. Euphytica, vol. 213, p. 103. https://doi.org/10.1007/s10681-017-1882-8
Chichkova, N.V.; Galiullina, R.A.; Mochalova, L.V.; Trusova, S.V.; Sobri, Z.M., Gallois, P. and Vartapetian, A. (2017) - Arabidopsis thaliana phytaspase: identification and peculiar properties. Functional Plant Biology, vol. 45, n. 2, p. 171-179. https://doi.org/10.1071/FP16321 [ Links ]
Chichkova, N.V.; Shaw, J.; Galiullina, R.A.; Drury, G.E.; Tuzhikov, A.I.; Kim, S.H.; Kalkum, M.; Hong, T.B.; Gorshkova, E.N.; Torrance, L.; Vartapetian, A. & Taliansky, M. (2010) - Phytaspase, a relocalisable cell death promoting plant protease with caspase specificity. The EMBO Journal, vol. 29, n. 6, p. 1149–1161. https://doi.org/10.1038/emboj.2010.1 [ Links ]
Dodson, G. and Wlodawer, A. (1998) - Catalytic triads and their relatives. Trends in Biochemical Sciences, vol. 23, n. 9, p. 347–352. https://doi.org/10.1016/S0968-0004(98)01254-7 [ Links ]
Duan, X.; Zhang, Z.; Wang, J. and Zuo, K. (2016) - Characterization of a novel cotton subtilase gene GbSBT1 in response to extracellular stimulations and its role in Verticillium resistance. PLoS One, vol. 11, art. e0153988. https://doi.org/10.1371/journal.pone.0153988 [ Links ]
Ekchaweng, K.; Khunjan, U. and Churngchow, N. (2017) - Molecular cloning and characterization of three novel subtilisin-like serine protease genes from Hevea brasiliensis. Physiological and Molecular Plant Pathology, vol. 97, p. 79–95. https://doi.org/10.1016/j.pmpp.2016.12.007. [ Links ]
Fan, T.; Bykova, N.V.; Rampitsch, C. and Xing, T. (2016) - Identification and characterization of a serine protease from wheat leaves. European Journal of Plant Pathology, vol. 146, n. 2, p. 293–304. https://doi.org/10.1007/s10658-016-0914-x [ Links ]
Figueiredo, J.; Costa, G.J.; Maia, M.; Paulo, O.S.; Malhó, R.; Sousa Silva, M. and Figueiredo, A. (2016) - Revisiting Vitis vinifera subtilase gene family: a possible role in grapevine resistance against Plasmopara viticola. Frontiers in Plant Science, vol. 7, art. 1783. https://doi.org/10.3389/fpls.2016.01783 [ Links ]
Figueiredo, J.; Sousa Silva, M. and Figueiredo, A. (2018) - Subtilisin-like proteases in plant defence: the past, the present and beyond. Molecular Plant Pathology, vol. 19, n. 4, p. 1017–1028. https://doi.org/10.1111/mpp.12567 [ Links ]
Glazebrook, J. (2005) - Contrasting Mechanisms of Defense Against Biotrophic and Necrotrophic Pathogens. Annual Review of Phytopathology, vol. 43, p. 205–227. https://doi.org/10.1146/annurev.phyto.43.040204.135923 [ Links ]
Guerreiro, A.; Figueiredo, J.; Sousa Silva, M. and Figueiredo, A. (2016) - Linking jasmonic acid to grapevine resistance against the biotrophic oomycete Plasmopara viticola. Frontiers in Plant Science, vol. 7, art. 565. https://doi.org/10.3389/fpls.2016.00565 [ Links ]
Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F. and Vandesompele, J. (2007) - qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biology, vol. 8, art. R19. https://doi.org/10.1186/gb-2007-8-2-r19 [ Links ]
Li, Y. and Inouye, M. (1994) - Autoprocessing of prothiolsubtilisin E in which active-site serine 221 is altered to cysteine. Journal of Biological Chemistry, vol. 269, n. 6, p. 4169–4174. [ Links ]
Lindstrom-Lang, K, and Ottesen, M. (1947) - A new protein from ovalbumin. Nature, vol. 159, p. 807–808. https://doi.org/10.1038/159807a0 [ Links ]
Monteiro, F.; Sebastiana, M.; Pais, M.S. and Figueiredo, A. (2013) - Reference gene selection and validation for the early responses to downy mildew infection in susceptible and resistant Vitis vinifera cultivars. PLoS One, vol. 8, art. e72998. https://doi.org/10.1371/journal.pone.0072998 [ Links ]
OIV (2017) - Statistical Report on World Vitiviniculture. International Organisation of Vine and Wine (OIV). [ Links ]
Ramírez, V.; López, A.; Mauch-Mani, B.; Gil, M.J. and Vera, P. (2013) - An extracellular subtilase switch for immune priming in Arabidopsis. PLoS Pathogens, vol. 9, art. e1003445. https://doi.org/10.1371/journal.ppat.1003445 [ Links ]
Rawlings, N.D. and Salvesen, G. (2013) - Handbook of proteolytic enzymes. 3ª ed. Amsterdam: Elsevier/AP. [ Links ]
Salzman, R.A.; Brady, J.A.; Finlayson, S.A.; Buchanan, C.D.; Summer, E.J.; Sun, F.; Klein, P.E.; Klein, R.R.; Pratt, L.H.; Cordonnier-Pratt, M. and Mullet, J.E. (2005) - Transcriptional profiling of sorghum induced by methyl jasmonate, salicylic acid, and aminocyclopropane carboxylic acid reveals cooperative regulation and novel gene responses. Plant Physiology, vol. 138, p. 352–368. https://doi.org/10.1104/pp.104.058206 [ Links ]
Schaller, A. and Ryan, C.A. (1994) - Identification of a 50-kDa systemin-binding protein in tomato plasma membranes having Kex2p-like properties. Proceedings of the National Academy of Sciences of the USA, vol. 91, n. 25, p. 11802–11806. [ Links ]
Schaller, A.; Stintzi, A.; Rivas, S.; Serrano, I.; Chichkova, N.V.; Vartapetian, A.B.; Martínez, D.; Guiamét, J.J.; Sueldo, D.J.; van der Hoorn, R.A.L.; Ramírez, V. and Vera, P. (2018) - From structure to function – a family portrait of plant subtilases. New Phytologist, vol. 218, n. 3, p. 901-915. https://doi.org/10.1111/nph.14582 [ Links ]
Siezen, R.J. and Leunissen, J.A.M. (1997) - Subtilases: The superfamily of subtilisin-like serine proteases. Protein Science, vol. 6, n. 3, p. 501–523. https://doi.org/10.1002/pro.5560060301. [ Links ]
Sun, J.-Q.; Jiang, H.-L. and Li, C.-Y. (2011) - Systemin/Jasmonate-Mediated Systemic Defense Signaling in Tomato. Molecular Plant, vol. 4, n. 4, p. 607–615. https://doi.org/10.1093/mp/ssr008 [ Links ]
Tornero, P.; Conejero, V. and Vera, P. (1996) - Primary structure and expression of a pathogen-induced protease (PR-P69) in tomato plants: similarity of functional domains to subtilisin-like endoproteases. Proceedings of the National Academy of Sciences of the USA, vol 93, n. 13, p. 6332–6337. [ Links ]
Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A. and Speleman, F. (2002) - Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology, vol. 3, res. 0034.1. https://doi.org/10.1186/gb-2002-3-7-research0034 [ Links ]
Zhu, X.; Ohta, Y.; Jordan, F. and Inouye, M. (1989) - Pro-sequence of subtilisin can guide the refolding of denatured subtilisin in an intermolecular process. Nature, vol. 339, p. 483–484. https://doi.org/10.1038/339483a0 [ Links ]
Acknowledgments
This work was supported by projects PEst-OE/BIA/UI4046/2014, PEst-OE/QUI/UI0612/2013, UID/MULTI/00612/2013, investigator FCT program IF/00819/2015 and grant SFRH/BD/116900/2016 from Fundação para a Ciência e Tecnologia (FCT/MCTES/PIDDAC, Portugal) and Joana Figueiredo PhD grant from Universidade de Lisboa.
Received/recebido: 2017.12.15
Accepted/aceite: 2018.05.18
These authors are co-senior authors in this paper