Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Similares em
SciELO
Similares em
SciELO
Compartilhar
Revista de Ciências Agrárias
versão impressa ISSN 0871-018X
Rev. de Ciências Agrárias vol.42 no.4 Lisboa dez. 2019
https://doi.org/10.19084/rca.18577
ARTIGO
Molecular factors associated with pathogenicity of Phytophthora cinnamomi
Fatores moleculares associados com a patogenicidade de Phytophthora cinnamomi
Darling de Andrade Lourenço1, Priscila Marques Moura de Leon1, Luís Santos2 & Altino Branco Choupina3,*
1 Biotechnology Department/ Technological Development Center, Federal University of Pelotas, Campus Universitário, s/nº., 96010-900, Pelotas, RS, Brazil
2 Department of Production and Plant Technology of Agricultural College of Bragança, Polytechnic Institute of Bragança, Campus Santa Apolónia, 5301-855, Bragança, Portugal
3 CIMO-Mountain Research Center, Department of Biology and Biotechnology, Agricultural College of Bragança, Polytechnic Institute of Bragança, Campus Santa Apolónia, 5301-855, Bragança, Portugal
(*E-mail: albracho@ipb.pt)
ABSTRACT
Phytophthora cinnamomiis soil pathogen that has a wide range of hosts in several countries and different climates. This fungus is responsible for the chestnut ink disease (Castanea sativaMiller) and death of the tree. Portugal stands out in the production of the European chestnut tree. However, between 2002 and 2004, there was a decrease of 27.3% in the distribution area of this tree due to P. cinnamomi. The aim of this study was to identify molecular factors possibly associated with the fungal pathogenicity through genomic sequences deposited at NCBI using bioinformatics tools. The first contig was used and the OFRs present in de sequences were identified. SmartBlast was used for homologous proteins. The prediction of cellular localization prediction of proteins was performed using four different tools: SignalP 4.1, Cello v.2.5, LOCTree3, Euk-mPLoc 2.0. Protein domains characterization was accomplished using PROSITE and the structure prediction was performed using Phyre2 server. We found 13 proteins probably associated with the pathogenicity of P. cinnamomi and its properties related to the infection were analyzed in silico.These results are important since they are a first step in the search for pathogenic factors.
Keywords:Chestnut ink disease, Oomycetes, Infection, Bioinformatics.
RESUMO
Phytophthora cinnamomié um patógeno do solo que possui uma ampla gama de hospedeiros em diversos países de diferentes climas. Esse fungo é responsável pela doença da tinta do castanheiro europeu (Castanea sativaMiller) e conduz à morte da árvore. Portugal se destaca na produção do castanheiro europeu. Entretanto, entre 2002 e 2004, houve uma redução de 27.3% na área de distribuição dessa área devido ao P. cinnamomi. O objetivo deste trabalho foi identificar fatores moleculares possivelmente associados à patogenicidade do fungo através de sequências genômicas depositadas no NCBI utilizando ferramentas de bioinformática. O primeiro contig foi utilizado e as ORFs presentes nas sequências foram identificadas. Foi utilizado o smartBlast em busca de proteínas homólogas. A predição da localização celular das proteínas foi feita através de quatro ferramentas: SignalP 4.1, Cello v.2.5, LOCTree3, Euk-mPLoc 2.0. A caracterização dos domínios dos produtos foi realizada utilizando o PROSITE e a predição das estruturas foi realizada utilizando o servidor Phyre2. Foram encontradas 13 proteínas provavelmente associadas com a patogenicidade de P. cinnamomie as suas propriedades relacionadas com a infecção foram analisadas in silico.Esses resultados são importantes uma vez que são uma primeira etapa na busca por fatores patogênicos.
Palavras-chave:Tinta do castanheiro, Oomycetes, Infecção, Bioinformática.
INTRODUCTION
The Oomycete fungus Phytophthora cinnamomiis a soil pathogen extremely aggressive and it has a wide range of hosts, that includes almost 500 plants species in more than 70 countries (Jung et al., 2013). It was described firstly in Sumatra (Indonesia) by Rands (1922) as being the causative agent of cinnamon (Cinnamomum burmannii) stripe cancer.
Due to its potential pathogenicity, P. cinnamomi generates substantial economic losses in agriculture, afforestation, and horticulture. This pathogen is associated with the oak decline in Mediterranean Europe and with the oak disease in California (Brasier et al., 1993; Robin et al., 1998; Vettraino et al., 2002; Garbelotto and Hüberli, 2006). Further, it causes root rot in fynbos in South Africa, leading to its death (von Broembsen & Kruger, 1985; Nagel et al., 2013). In addition, its introduction in nature leads to severe consequences to the natural ecosystem and biodiversity (Hardham and Blackman, 2018).
The Oomycetes class, which includes genus Phytophthora, belongs to Pseudofungi phylum within the Chromista kingdom. The characteristics that define the Chromista are the asexual reproduction of mobile spores endowed with a flagellum adorned by tubular hairs, which are responsible for motility. These spores start the plant infection in many species of Phytophthora, including P. cinnamomi(Beakes et al., 2012; Hardham and Blackman, 2018). P. cinnamomialso is responsible for the European chestnut ink disease (Castanea sativaMiller) and, worsening this problem, C. sativa is very susceptible to infections by this pathogen (de Sampaio e Paiva Camilo-Alves et al., 2013). The symptoms of this disease include yellowing and premature fall off the leaves and the appearance of wet rot in the roots, which will lead to the death of the tree (Fernandes, 1953; Shattock, 2001).
The European chestnut tree has economic interests due to the fruit and wood production, as well as other Castaneaspecies. Portugal is the third-largest European chestnut tree producer, with 27.337 tons in 35.426 hectares (INE, 2016) and the Cold Land Chestnut Production Zone is responsible for the production of 80% of the national chestnut (INIAV, 2016). However, the C. sativaproduction has been declining over the years due to the epidemic proportions of P. cinnamomi, only between 2002 and 2004, the area of distribution decreased 27.3 % (Martins et al., 2007; Pereira, 2017).
Despite several species are affected by P. cinnamomi, European chestnut trees receive special highlights in Portugal due to the economic importance for the country (INIAV, 2016). Additionally, P. cinnamomi is ranked as one of the most aggressive species of Phytophthora genus to the C. sativa (Vettraino et al., 2001). However, the ink disease still the main threat once P. cinnamomi is resistant to the available fungicides and there are no effective options of pest control (Santos et al., 2016). Still, the molecular mechanisms involved in the pathogenicity of P. cinnamomi remains unclear.
The plant infection by P. cinnamomiis initiated by binding the zoospores to the plant root elongation region (Hardham, 2005). This binding is provided by some proteins, like adhesine Vsv1 (Hardham and Gubler, 1990; Robold and Hardham, 2005). Then, zoospores enter the stage of encystment with the formation of biofilm from secreted proteins. Between 20-30 minutes after the zoospores encystment, the cyst germinates and the hyphae are formed (Hua et al., 2013; Hardham and Blackman, 2018). These hyphae are responsible for the production of some enzymes that will degrade the plant cell wall, such as the ones that belong to the Cell Wall-Degrading Enzymes (CWDEs) family (Götesson et al., 2002; Ospina-Giraldo et al., 2010; Larroque et al., 2012; Blackman et al., 2014). The intracellular and intercellular growth of hyphae in the radicular cortex towards the cortical and vascular tissues leads to water stress and necrosis (Hosseini, 2010; Hardham and Blackman, 2018).
Bioinformatic provide promising tools for analysis in silico and to support in vivo assays. In the context of this work, ORFFinder and smartBLAST are two tools from NCBI that enable an easy and trustworthy search for open read frames in genomic sequences and provides determination of homology of these sequences (Rawat et al., 2018). Concerning the protein structures, the SignalP 4.1 is a reliable tool for the prediction of the signal peptide (Petersen et al., 2011; Ayalew et al., 2017). Considering that secreted proteins are highly important to the pathogenicity of pathogens, the prediction of subcellular localization is indispensable. Among the available online bioinformatics tools LocTree3 (Nair and Rost, 2005; Sanasam and Kumar, 2019), Cello (Yu et al., 2004; Thakare et al., 2016) and Euk-mPLoc (Chou, 2019) stand out. In line with this, the identification of domains and motifs of proteins are very useful to determine the proteins functions. PROSITE is an online tool for searching for significant sites in amino acid sequences (Gellért et al., 2006; Sigrist et al., 2009). Finally, determine the protein structure through modeling is essential to analyze its conformation. For this use, PHYRE2 is a very used web-based tool for modeling amino acid sequences (Zhang et al., 2011; Singh and Gupta, 2016).
Considering the economic losses caused by P. cinnamomiin European chestnut trees it is extremely important that molecular factors responsible for the pathogenicity of this pathogen are identified. Thus, the aim of this study was to identify genes and its products related to the pathogenicity of P. cinnamomi, as well as the characterization of the results obtained through bioinformatics tools.
MATERIAL AND METHODS
Database of biological information
For the search of genes, the genomic sequences from Phytophthora cinnamomiused are deposited in the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/). To continue the research from the group used the sequence (REF: LGSK01000001.1) deposited by Studholme et al. (2016) which belongs to the strain MP94-48 (BioSample: SAMN03921829; BioProject: PRJNA290836; Assembly: GCA_001314365.1).
Open Read Frames (ORFs) search and homology
In order to find de ORFs present in genomic sequences we used the ORFFinder tool (https://www.ncbi.nlm.nih.gov/orffinder/), available by NCBI, with the following parameters: minimal ORF length: 300 nucleotides; genetic code: Standard; ORF start codon to use: ATG only.
The homology of proteins encoded by the ORFs founded was determined using the Basic Local Alignment Search Tool (Blast), specifically the smartBlast version, also available by NCBI. This tool establishes the homology between the sequences deposited in several protein databases available.
Prediction of cellular localization of proteins
The software SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/) was used to analyze the presence of signal-peptide. The following softwares were used to predict the sub-cellular location of proteins: Cello v.2.5 (http://cello.life.nctu.edu.tw/) ; LOCTree3 (https://rostlab.org/services/loctree3/); and Euk-mPLoc 2.0 (http://www.csbio.sjtu.edu.cn/bioinf/euk-multi-2/). Each one analyzes proteins in different ways and for all tools, the selected parameters were: "Eukaryotes" for organism or domain and "Protein" for the type of sequence.
Gene and proteins characterization
Genes and their products were characterized through bioinformatics tools that allow the identification of domains, patterns, and motifs, as well as active center and physical-chemical characteristics. Therefore, we used the PROSITE (https://prosite.expasy.org/) tool, in Quick Scan mode, excluding motifs with a high probability of occurrence.
The structural prediction of the proteins found was performed using the Phyre2 (http://www.sbg.bio.ic.ac.uk/~phyre2/html/page.cgi?id=index) server, using the normal modeling mode. For visualization and representation of the structures, we used the Jmol program.
RESULTS AND DISCUSSION
1980 ORFs, larger than 300 nucleotides, established by the ORFfinder program, were analyzed using the smartBlast in search of homology. Of these ORFs, we found 13 ORFs that encoded for proteins homologous related to the pathogenicity in different organisms mostly of the genus Phytophthora, as shown in Table 1. The selection method was based on a link between the protein sequence and the pathogenicity in other organisms. This link was established with references in the scientific literature available. The data about the genes chosen and their related products are presented in Table 1. All information about these genes and their products including scientific references can be obtained from the databases referred to in Table 1 through the accession number of each gene in the respective database.
The prediction of cellular localization of proteins informs their destination after leaving the endoplasmic reticulum. This destination often depends on the signaling of a signal peptide. The results of the prediction of cellular localization of the proteins encoded by the ORFs selected are in Table 2. To obtain these results we used: SignalP to determine the presence or absence of signal peptide in the protein; Cello, LocTree3, and Euk-mPloc, which from different algorithms, predict the cellular localization of proteins.
The results obtained with the three prediction tools diverge sometimes due to the different algorithms that they use. In addition, some of these proteins are not determined molecularly by experiments in vitro, being determined only in silico by bioinformatics methods, a factor that hinders the prediction by the tools. According to Dönnes and Höglund (2004), the several web-based methods for the prediction that are available online have different localization coverage and different means to assess their accuracy, making it impossible to compare all methods against each other. Some examples of this divergence can be seen in Table 2 where it can also be observed that most proteins with a role in pathogenicity have extracellular or membrane fate as expected as it is outside the parasite cell and in interaction with host cells that they will exercise their action.
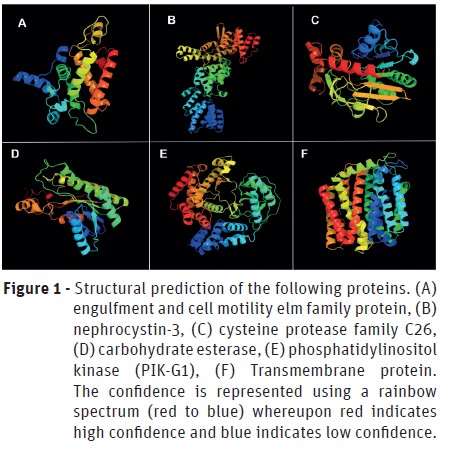
The characterization of genes and its products was performed using Phyre2 server for modeling and structure prediction. The domains and active sites were identified using PROSITE tool. The structures predicted by modeling are in Figure 1 and 2 and in Table 3 is possible to analyze the domains and active sites detected. However, the structure of E3 ubiquitin-protein ligase HERC2 could not be predicted since it has a very long amino acid chain.
The most promising protein found was the crinkler (CRN) family. This protein is widely related to the damage caused by the Phytophthora genus in its hosts (Lamour et al., 2007; Fawke et al., 2015). It was first identified in Phytophthora infestans but is known to be present in almost all species of plant pathogenic oomycetes (Torto et al., 2003). It is known that CRN family interacts with the host DNA, leading to leaf wrinkles and necrosis (Amaro et al., 2017). The mature CRNs are able to induce cell death through translocation of cytoplasmatic factors and also by action on the host nucleus (Schornack et al., 2010; Stam et al., 2013). This protein family probably is present at the base of the late stages of Phytophthora cinnamomiinfection due to its functioning.
Carbohydrate esterase is an enzyme that catalyzes the reaction of hydrolysis in ester bonds in order to remove oxygen (O) or nitrogen (N) from carbohydrates (Cantarel et al., 2009). Although the sequence found does not have any specific domain found, it is still possible to perceive its probable importance in P. cinnamomiinfection, through the breakdown of complex carbohydrates and glycoconjugates of the host, such as cellulose from cells wall. Cysteine protease family C26 is a protein with peptidase activity and involved in the metabolic process of glutamine due to its GATASE_TYPE_1 (Glutamine amidotransferase type 1) domain. This domain is responsible for catalyzing the reaction of removal of the ammonia group from glutamine and transfer to another substrate to form a new carbon-nitrogen group (Buchanan, 1973). Cysteine proteases are conserved among the animal and plant pathogenic microorganisms, besides proteases facilitate penetration of the plant hosts physical barriers.
Additionally, both cysteine protease and papain-like cysteine protease C1 are enzymes with the proteolytic activity of the family eukaryotic thiol proteases, which have cysteine in their active sites (Dufour, 1988). In addition to the proteolytic action, both also have the domain of the family phosphofructokinase B-type of carbohydrate kinases, which has a function not yet well defined, however, it seems to be involved with the plastidial genome (Yokota et al., 2009). Thus, due to the activity of these protein, it is possible to assume that they have an influence on the pathogenicity of P. cinnamomisince it degrades peptides through their proteolytic actions, facilitating the pathogen penetration into the hosts tissues.
In line with this, serine protease has two carboxypeptidase active sites: serine/serine and serine/histidine, involving an aspartic acid residue in the catalysis system (Liao and Remington, 1990). Carboxypeptidases have proteolytic action and cleave near the carboxyl end site (Beekman et al., 2018). Several studies already associate the presence of carboxypeptidases with pathogenicity fungi, such as Pseudogymnoascus destructans (Beekman et al., 2018), and the bacteria of the genus Neisseriasp. (Schaub & Dillard, 2019).
E3 ubiquitin-protein ligase HERC4 and E3 ubiquitin-protein ligase HERC2 have the B30.2_SPRY domain, which is well-conserved and helps to regulate spore differentiation (Nuckolls et al., 1996). Considering the importance of the spores in the infection of C. sativa by P. cinnamomi is possible that these enzymes are related to the pathogenicity of the present pathogen in the early stages. Glycoside hydrolase has hydrolase and transferase activity, i.e., has the ability to cleave glycosidic bonds as well as assist in cell wall elongation (Haas et al., 2009). It has two distinct domains, with one active site in each. The action of this enzyme in the pathogenicity of P. cinnamomiis possibly originated in its lysing activity in glycosidic bonds, altering the permeability of the host.
As less promising proteins, yet very important, we found that the engulfment and cell motility elm family protein (Figure 1 A) belongs to the ELMO (Engulfment and Cell Motility) family of ELMO and PH domains. The ELMO domain is responsible for the regulatory activity of GTPases (Bowzard et al., 2007), whereas the PH (Pleckstrin homology) domain occurs in several proteins that perform intracellular signaling or constitute the cytoskeleton (Haslam et al., 1993; Mayer et al., 1993; Musacchio et al., 1993; Gibson et al., 1994; Ingley and Hemmings, 1994; Pawson, 1995; Saraste and Hyvönen, 1995). Besides that, it is known that some pathogens, such as Shigella flexneri, promotes bacterial entry in cells through the ELMO-Dock180 machinery (Hachani et al., 2008). Due to its importance in cell motility and the evidences involving the ELMO domain with pathogenicity, it is possible to infer that this protein participates in the infectious process by P. cinnamomiin the initial stages utilizing some homolog protein to Dock180.
The nephrocystin-3 is a protein involved in the ciliary development and function and thus, involved in cell movement (Bergmann et al., 2008). It has the SAM domain (Sterile α Motif), which is related to protein-protein interaction (Thanos et al., 1999), as well as the TPR (Tetratrico Peptide Repeat) domain, which is responsible for several processes, such as cell cycle control, stress response, transcription repression, among others (Lamb et al., 1995). TPR domain is present in some proteins that are directly involved in bacterial virulence-associated functions in animal cells and tissue. These functions include the translocation of virulence factors into host cells and adhesion to host cells (Edqvist et al., 2006; Chakraborty et al., 2008; Chao et al., 2010). Due to the activities performed by the domains of nephrocystin-3, it is possible to infer that it has high importance in pathogenicity and cellular movements and development – both of these processes, being essential for the infection by P. cinnamomi.
Phosphatidylinositol kinase (PIK-G1) is an enzyme responsible for the molecular function of the kinase (transference of phosphate groups) and complexation of metal ions. Due to this, it is involved in the biological processes of phosphatidylinositol phosphorylation and signaling (Kaur et al., 2010). It has PH domain, reinforcing its function in intracellular signaling and, the PI3_4_KINASE_3 (Phosphatidylinositol 3- and 4-kinases) domain, involved in cell signaling of several metabolic pathways and cell cycle (Hiles et al., 1992; Haslam et al., 1993; Mayer et al., 1993; Musacchio et al., 1993; Ingley and Hemmings, 1994; Pawson, 1995; Saraste and Hyvönen, 1995). In line with this, PIK-G1 must play a key role in the stage of cell proliferation during the infectious process.
Although the transmembrane proteins do not integrate any metabolic pathway, they have already been reported in the pathogenicity of several microorganisms, since it is through them that effector molecules and toxins can be released to the host (Silveira et al., 2016). Said that it is important to highlight the relevance of the transmembrane proteins in the process of infection by P. cinnamomithrough the release of pathogenic effector molecules produced intracellularly.
CONCLUSIONS
1. Bioinformatic tools are a reliable source to perform prediction ins proteins sequences available in databases while in vitrostudies still not performed, and also as a previous step to better target these experimental studies.
2. New research using P. cinnamomigenomic sequence must be carried out in order to determine its pathogenicity mechanisms and molecules associated, and possibly develop methods of control of the pathogens and reduce the damage to European chestnut tree.
3. It is possible to deduce the role of many molecules in the metabolic pathways by their similitude and homology, however, there is the possibility that many molecules have roles that are still unknown today.
4. As demonstrated by (Pascoal et al., 2018), we also believe that the silencing of these genes in phytopathogenic fungi and the subsequent infection of plants by these fungi may help to better understand the role of the enzymes expressed by these genes in the infection mechanism.. The research for molecular factors as well as the metabolic pathways involved in the infectious processes is an important tool for the development of preventive strategies and control of pathogens in plants.
REFERENCES
Amaro, T.M.M.M.; Gaëtan, J.A.; Thilliez, G.B.M. & Huitema, E. (2017) - A Perspective on CRN Proteins in the Genomics Age: Evolution, Classification, Delivery and Function Revisited. Frontiers in Plant Science, vol. 8, art. 99. https://doi.org/10.3389/fpls.2017.00099 [ Links ]
Ayalew, S.; Confer, A.W.; Hartson, S.D.; Canaan, P.J.; Payton, M. & Couger, B. (2017) - Proteomic and Bioinformatic Analyses of Putative Mannheimia haemolytica Secretome by Liquid Chromatography and Tandem Mass Spectrometry. Veterinary Microbiology, vol. 203, p. 73–80. https://doi.org/10.1016/j.vetmic.2017.02.011 [ Links ]
Beakes, G.W.; Glockling, S.L. & Sekimoto, S. (2012) - The Evolutionary Phylogeny of the Oomycete Fungi. Protoplasma, vol. 249, n. 1, p. 3–19. https://doi.org/10.1007/s00709-011-0269-2 [ Links ]
Beekman, C.; Jiang, Z.; Suzuki, B.M.; Palmer, J.M.; Lindner, D.L.; ODonoghue, A.J.; Knudsen, G.M. & Bennett, R.J. (2018) - Characterization of PdCP1, a Serine Carboxypeptidase from Pseudogymnoascus destructans, the Causal Agent of White-Nose Syndrome. Biological Chemistry, vol. 399, n. 12, p. 1375–1388. https://doi.org/10.1515/hsz-2018-0240 [ Links ]
Bergmann, C.; Fliegauf, M.; Brüchle, N.O.; Frank, V.; Olbrich, H.; Kirschner, J.; Schermer, B.; Schmedding, I.; Kispert, A.; Kränzlin, B.; Nürnberg, G.; Becker, C.; Grimm, T.; Girschick, G.; Lynch, S.A.; Kelehan, P.; Senderek, J.; Neuhaus, T.J.; Stallmach, T.; Zentgraf, H.; Nürnberg, P.; Gretz, N.; Lo, C.; Lienkamp, S.; Schäfer, T.; Walz, G.; Benzing, T.; Zerres, K. & Omran, H. (2008) - Loss of Nephrocystin-3 Function Can Cause Embryonic Lethality, Meckel-Gruber-like Syndrome, Situs Inversus, and Renal-Hepatic-Pancreatic Dysplasia. The American Journal of Human Genetics, vol. 82, n. 4, p. 959–970. https://doi.org/10.1016/j.ajhg.2008.02.017 [ Links ]
Blackman, L.M.; Cullerne, D.P. & Hardham, A.R. (2014) - Bioinformatic Characterisation of Genes Encoding Cell Wall Degrading Enzymes in the Phytophthora parasitica Genome. BMC Genomics, vol. 15, n. 1, art. 785. https://doi.org/10.1186/1471-2164-15-785 [ Links ]
Bowzard, J.B.; Cheng, D.; Peng, J. & Kahn, R.A. (2007) - ELMOD2 Is an Arl2 GTPase-Activating Protein That Also Acts on Arfs. Journal of Biological Chemistry, vol. 282, n. 24, p. 17568–17580. https://doi.org/10.1074/jbc.M701347200 [ Links ]
Brasier, C.M., Robredo, F. & Ferraz, J.F.P. (1993) - Evidence for Phytophthora cinnamomi Involvement in Iberian Oak Decline. Plant Pathology, vol. 42, n. 1, p. 140–245. https://doi.org/10.1111/j.1365-3059.1993.tb01482.x [ Links ]
von Broembsen, S. L.; Kruger, F. J. (1985). Phytophthora cinnamomiassociated with mortality of native vegetation in South Africa. Plant Disease, vol. 69, p. 715-717. [ Links ]
Buchanan, J.M. (1973) - The Amidotransferases. In: Advances in Enzymology and Related Areas of Molecular Biology, vol. 39, p. 91–183. [ Links ]
Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V. & Henrissat, B. (2009) - The Carbohydrate-Active EnZymes Database (CAZy): An Expert Resource for Glycogenomics. Nucleic Acids Research, vol. 37, p. D233–D238. https://doi.org/10.1093/nar/gkn663 [ Links ]
Chakraborty, S.; Monfett, M.; Maier, T.M.; Benach, J.L.; Frank, D.W. and Thanassi, D.G. (2008) - Type IV Pili in Francisella tularensis: Roles of PilF and PilT in Fiber Assembly, Host Cell Adherence, and Virulence. Infection and Immunity, vol. 76, n. 7, p. 2852–2861. https://doi.org/10.1128/IAI.01726-07 [ Links ]
Chao, J.; Wong, D.; Zheng, X.; Poirier, V.; Bach, H.; Hmama, Z. & Av-Gay, Y. (2010) - Protein Kinase and Phosphatase Signaling in Mycobacterium tuberculosis Physiology and Pathogenesis. Biochimica et Biophysica Acta, vol. 1804, n. 3, p. 620–627. https://doi.org/10.1016/j.bbapap.2009.09.008 [ Links ]
Chou, K.-C. (2019) - An Insightful Recollection for Predicting Protein Subcellular Locations in Multi-Label Systems. Genomics, in press. https://doi.org/10.1016/j.ygeno.2019.08.008 [ Links ]
Dönnes, P. & Höglund, A. (2004) - Predicting Protein Subcellular Localization: Past, Present, and Future. Genomics, Proteomics & Bioinformatics, vol. 2, n. 4, p. 209–215. https://doi.org/10.1016/S1672-0229(04)02027-3 [ Links ]
Dufour, E. (1988) - Sequence Homologies, Hydrophobic Profiles and Secondary Structures of Cathepsins B, H and L: Comparison with Papain and Actinidin. Biochimie, vol. 70, n. 10, p. 1335–1342. https://doi.org/10.1016/0300-9084(88)90004-1 [ Links ]
Edqvist, P.J.; Broms, J.E.; Betts, H.J.; Forsberg, A.; Pallen, M.J. & Francis, M.S. (2006) - Tetratricopeptide Repeats in the Type III Secretion Chaperone, LcrH: Their Role in Substrate Binding and Secretion. Molecular Microbiology, vol. 59, n. 1, p. 31–44. https://doi.org/10.1111/j.1365-2958.2005.04923.x [ Links ]
Fawke, S.; Doumane, M. & Schornack, S. (2015) - Oomycete Interactions with Plants: Infection Strategies and Resistance Principles. Microbiology and Molecular Biology Reviews, vol. 79, n. 3, p. 263–280. https://doi.org/10.1128/MMBR.00010-15 [ Links ]
Fernandes, C.T. (1953) - A Luta Contra a Doença Da Tinta Dos Castanheiros No Norte de Portugal. Direção Geral Dos Serviços Florestais e Aquícolas, vol. 20, n. 2, p. 153–158. [ Links ]
Garbelotto, M. & Hüberli, D. (2006) - First Report on an Infestation of Phytophthora Cinnamomi in Natural Oak Woodlands of California and Its Differential Impact on Two Native Oak Species. Plant Disease, vol. 90, n. 5, p. 685. https://doi.org/10.1094/PD-90-0685C [ Links ]
Gellért, Á.; Salánki, K.; Náray-Szabó, G. & Balázs, E. (2006) - Homology Modelling and Protein Structure Based Functional Analysis of Five Cucumovirus Coat Proteins. Journal of Molecular Graphics and Modelling, vol. 24, n. 5, p. 319–327. https://doi.org/10.1016/j.jmgm.2005.09.015 [ Links ]
Gibson, T.J.; Hyvönen, M.; Musacchio, A.; Saraste, M. & Birney, E. (1994) - PH Domain: The First Anniversary. Trends in Biochemical Sciences, vol. 19, n. 9, p. 349–353. https://doi.org/10.1016/0968-0004(94)90108-2 [ Links ]
Götesson, A.; Marshall, J.S.; Jones, D.A. & Hardham, A.R. (2002) - Characterization and Evolutionary Analysis of a Large Polygalacturonase Gene Family in the Oomycete Plant Pathogen Phytophthora cinnamomi. Molecular Plant-Microbe Interactions, vol. 15, n. 9, p. 907–921. https://doi.org/10.1094/MPMI.2002.15.9.907 [ Links ]
Hachani, A.; Biskri, L.; Rossi, G.; Marty, A.; Ménard, R.; Sansonetti, Ph.; Parsot, C.; van Nhieu, G.T.; Bernardini, M.L. & Allaoui, A. (2008) - IpgB1 and IpgB2, Two Homologous Effectors Secreted via the Mxi-Spa Type III Secretion Apparatus, Cooperate to Mediate Polarized Cell Invasion and Inflammatory Potential of Shigella Flexenri. Microbes and Infection, vol. 10, n. 3, p. 260–268. https://doi.org/10.1016/j.micinf.2007.11.011 [ Links ]
Hardham, A. & Gubler, F. (1990) - Polarity of Attachment of Zoospores of a Root Pathogen and Pre-Alignment of the Emerging Germ Tube. Cell Biology International Reports, vol. 14, n. 11, p. 947–956. https://doi.org/10.1016/0309-1651(90)90107-A [ Links ]
Hardham, A.R. (2005) - Phytophthora cinnamomi. Molecular Plant Pathology, vol. 6, n. 6., p. 589–604. https://doi.org/10.1111/j.1364-3703.2005.00308.x [ Links ]
Hardham, A.R. & Blackman, L.M. (2018) - Phytophthora cinnamomi. Molecular Plant Pathology, vol. 19, n. 2, p. 260–285. https://doi.org/10.1111/mpp.12568 [ Links ]
Haslam, R.J.; Koide, H.B. & Hemmings, B.A. (1993) - Pleckstrin Domain Homology. Nature, vol. 363, n. 6427, p. 309–310. https://doi.org/10.1038/363309b0 [ Links ]
Haas, B.J., Kamoun, S., Zody, M.C., Jiang, R.H.Y., Handsaker, R.E., Cano, L.M., Grabherr, M., Kodira, C.D., Raffaele, S., Torto-Alalibo, T., Bozkurt, T.O., Ah- Fong, A.M.V., Alvarado, L., Anderson, V.L., Armstrong, M.R., Avrova, A., Baxter, L., Beynon, J., Boevink, P.C., Bollmann, S.R., Bos, J.I.B., Bulone, V., Cai, G.H., Cakir, C., Carrington, J.C., Chawner, M., Conti, L., Costanzo, S., Ewan, R., Fahlgren, N., Fischbach, M.A., Fugelstad, J., Gilroy, E.M., Gnerre, S., Green, P.J., Grenville-Briggs, L.J., Griffith, J., Grunwald, N.J., Horn, K., Horner, N.R., Hu, C.H., Huitema, E., Jeong, D.H., Jones, A.M.E., Jones, J.D.G., Jones, R.W., Karlsson, E.K., Kunjeti, S.G., Lamour, K., Liu, Z.Y., Ma, L.J., Maclean, D., Chibucos, M.C., McDonald, H., McWalters, J., Meijer, H.J.G., Morgan, W., Morris, P.F., Munro, C.A., ONeill, K., Ospina-Giraldo, M., Pinzon, A., Pritchard, L., Ramsahoye, B., Ren, Q.H., Restrepo, S., Roy, S., Sadanandom, A., Savidor, A., Schornack, S., Schwartz, D.C., Schumann, U.D., Schwessinger, B., Seyer, L., Sharpe, T., Silvar, C., Song, J., Studholme, D.J., Sykes, S., Thines, M., van de Vondervoort, P.J.I., Phuntumart, V., Wawra, S., Weide, R., Win, J., Young, C., Zhou, S.G., Fry, W., Meyers, B.C., Van West, P., Ristaino, J., Govers, F., Birch, P.R.J., Whisson, S.C., Judelson, H.S. and Nusbaum, C. (2009) Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature, vol. 461, p. 393–398. [ Links ]
Hiles, I.D.; Otsu, M.; Volinia, S.; Fry, M.J.; Gout, I.; Dhand, R.; Panayotou, G.; Ruiz-Larrea, F.; Thompson, A. & Totty, N.F. (1992) - Phosphatidylinositol 3-Kinase: Structure and Expression of the 110 Kd Catalytic Subunit. Cell, vol. 70, n. 3, p. 419–429. https://doi.org/10.1016/0092-8674(92)90166-A [ Links ]
Hosseini, S. (2010) - Expression Patterns of Pathogenicity Genes during Phytophthora pisi Infection of Pea Roots. Plant Biology-Masters Programme, Second cycle, A1E. Uppsala: SLU, Dept. of Forest Mycology and Plant Pathology. [ Links ]
Hua, C.; Meijer, H.J.G.; de Keijzer, J.; Zhao, W.; Wang, Y. & Govers, F. (2013) - GK4, a G-Protein-Coupled Receptor with a Phosphatidylinositol Phosphate Kinase Domain in Phytophthora infestans, Is Involved in Sporangia Development and Virulence. Molecular Microbiology, vol. 88, n. 2, p. 352–370. https://doi.org/10.1111/mmi.12190 [ Links ]
INE (2016) - Boletim Mensal Da Agricultura e Pescas - Dezembro 2016. Instituto Nacional de Estatística, Portugal. [ Links ]
Ingley, E. & Hemmings, B.A. (1994) - Pleckstrin Homology (PH) Domains in Signal Transduction. Journal of Cellular Biochemistry, vol. 56, n. 4, p. 436–443. https://doi.org/10.1002/jcb.240560403 [ Links ]
INIAV (2016) - Estudo Económico de Desenvolvimento Da Fileira Da Castanha Relatório Global. INIAV, Sabugal. [ Links ]
Jung, T.; Colquhoun, I. J. & Hardy, G. E. St. J. (2013) - New Insights into the Survival Strategy of the Invasive Soilborne Pathogen Phytophthora cinnamomi in Different Natural Ecosystems in Western Australia. Forest Pathology, vol. 43, n. 4, p. 266–88. https://doi.org/10.1111/efp.12025 [ Links ]
Kaur, J.; Sawhney, M.; DattaGupta, S.; Shukla, N.K.; Srivastava, A. & Ralhan, R. (2010) - Clinical Significance of Phosphatidyl Inositol Synthase Overexpression in Oral Cancer. BMC Cancer, vol. 10, art. 168. https://doi.org/10.1186/1471-2407-10-168 [ Links ]
Lamb, J.R.; Tugendreich, S. & Hieter, P. (1995) - Tetratrico Peptide Repeat Interactions: To TPR or Not to TPR? Trends in Biochemical Sciences, vol. 20, n. 7, p. 257–259. https://doi.org/10.1016/s0968-0004(00)89037-4 [ Links ]
Lamour, K.H.; Win, J. & Kamoun, S. (2007) - Oomycete Genomics: New Insights and Future Directions. FEMS Microbiology Letters, vol. 274, n. 1, p. 1–8. https://doi.org/10.1111/j.1574-6968.2007.00786.x [ Links ]
Larroque, M.; Barriot, R.; Bottin, A.; Barre, A.; Rougé, P.; Dumas, B. & Gaulin, E. (2012) - The Unique Architecture and Function of Cellulose-Interacting Proteins in Oomycetes Revealed by Genomic and Structural Analyses. BMC Genomics, vol. 13, n. 1, art. 605. https://doi.org/10.1186/1471-2164-13-605 [ Links ]
Liao, D.I. & Remington, S.J. (1990) - Structure of Wheat Serine Carboxypeptidase II at 3.5-A Resolution. A New Class of Serine Proteinase. The Journal of Biological Chemistry, vol. 265, n. 12, p. 6528–3651. https://doi.org/10.2210/pdb2sc2/pdb [ Links ]
Martins, L.; Castro, J.; Macedo, W.; Marques, C. & Abreu, C. (2007) - Assessment of the Spread of Chestnut Ink Disease Using Remote Sensing and Geostatistical Methods. European Journal of Plant Pathology, vol. 119, n. 2, p. 159–164. https://doi.org/10.1007/s10658-007-9155-3 [ Links ]
Mayer, B.J.; Ren, R.; Clark, K.L. & Baltimore, D. (1993) - A Putative Modular Domain Present in Diverse Signaling Proteins. Cell, vol. 73, n. 4, p. 629–630. https://doi.org/10.1016/0092-8674(93)90244-k [ Links ]
Musacchio, A.; Gibson, T.; Rice, P.; Thompson, J. & Saraste, M. (1993) - The PH Domain: A Common Piece in the Structural Patchwork of Signalling Proteins. Trends in Biochemical Sciences, vol. 18, n. 9, p. 343–348. https://doi.org/10.1016/0968-0004(93)90071-t [ Links ]
Nagel, J.H.; Gryzenhout, M.; Slippers, B. & Wingfield, M.J. (2013) - The Occurrence and Impact of Phytophthora on the African Continent. In: Phytophthora: a global perspective. Wallingford: CABI, p. 204–214. [ Links ]
Nair, R. & Rost, B. (2005) - Mimicking Cellular Sorting Improves Prediction of Subcellular Localization. Journal of Molecular Biology, vol. 348, n. 1, p. 85–100. https://doi.org/10.1016/j.jmb.2005.02.025 [ Links ]
Nuckolls, G.H.; Osherov, N.; Loomis, W.F. & Spudich, J.A. (1996) - The Dictyostelium Dual-Specificity Kinase SplA Is Essential for Spore Differentiation. Development, vol. 122, n. 10, p. 3295–3305. [ Links ]
Ospina-Giraldo, M.D.; Griffith, J.G.; Laird, E.W. & Mingora, C. (2010) - The CAZyome of Phytophthora spp.: A Comprehensive Analysis of the Gene Complement Coding for Carbohydrate-Active Enzymes in Species of the Genus Phytophthora. BMC Genomics, vol. 11, n. 1, art. 525. https://doi.org/10.1186/1471-2164-11-525 [ Links ]
Pascoal, A.; Estevinho, L.M.; Martins, I.M. & Choupina, A.B (2018) - Novel Sources and Functions of Microbial Lipases and Their Role in the Infection Mechanisms. Physiological and Molecular Plant Pathology, vol. 104, p. 119–126. https://doi.org/10.1016/j.pmpp.2018.08.003 [ Links ]
Pawson, T. (1995) - Protein Modules and Signalling Networks. Nature, vol. 373, n. 6515, p. 573–580. https://doi.org/10.1038/373573a0 [ Links ]
Pereira, N.A.F.N. (2017) - Caracterização de Fatores Moleculares Implicados Na Patogenicidade de Phytophthora cinnamomi. Dissertação de Mestrado, Instituto Politécnico de Bragança. [ Links ]
Petersen, T.N.; Brunak, S.; von Heijne, G. & Nielsen, H. (2011) - SignalP 4.0: Discriminating Signal Peptides from Transmembrane Regions. Nature Methods, vol. 8, n. 10, p. 785–786. https://doi.org/10.1038/nmeth.1701 [ Links ]
Rands, R.D. (1922) - Streepkanker van Kaneel, Veroorzaakt Door Phytophthora Cinnamomi n. sp. Batavia: Ruygrok. [ Links ]
Rawat, N.; Shanker, A. & Joshi, G.K. (2018) - Bioinformatics Analysis of a Novel Glutaredoxin Gene Segment from a Hot Spring Metagenomic DNA Library. Meta Gene, vol. 18, p. 107–111. https://doi.org/10.1016/j.mgene.2018.08.007 [ Links ]
Robin, C.; Desprez-Loustau, M.-L.; Capron, G. & Delatour, C. (1998) - First Record of Phytophthora cinnamomi on Cork and Holm Oaks in France and Evidence of Pathogenicity. Annales des Sciences Forestières, vol. 55, n. 8, p. 869–883. https://doi.org/10.1051/forest:19980801 [ Links ]
Robold, A.V. & Hardham, A.R. (2005) - During Attachment Phytophthora Spores Secrete Proteins Containing Thrombospondin Type 1 Repeats. Current Genetics, vol. 47, n. 5, p. 307–315. https://doi.org/10.1007/s00294-004-0559-8 [ Links ]
de Sampaio e Paiva Camilo-Alves, C.; da Clara, M.I.E. & de Almeida Ribeiro, N.M.C. (2013) - Decline of Mediterranean Oak Trees and Its Association with Phytophthora cinnamomi: A Review. European Journal of Forest Research, vol. 132, n. 3, p. 411–432. https://doi.org/10.1007/s10342-013-0688-z [ Links ]
Sanasam, B.D. & Kumar, S. (2019) - PRE-Binding Protein of Plasmodium falciparum Is a Potential Candidate for Vaccine Design and Development: An in Silico Evaluation of the Hypothesis. Medical Hypotheses, vol. 125, p. 119–123. https://doi.org/10.1016/j.mehy.2019.01.006 [ Links ]
Santos, C.; Machado, H.; Serrazina, S.; Gomes, F.; Gomes-Laranjo, J.; Correia, I.; Zhebentyayeva, T.; Duarte, S.; Bragança, H.; Fevereiro, P.; Dana Nelson, C. & Costa, R. (2016) - Comprehension of Resistance to Diseases in Chestnut. Revista de Ciências Agrárias, vol. 39, n. 2, p. 189–193. http://dx.doi.org/10.19084/RCA15145 [ Links ]
Saraste, M. & Hyvönen, M. (1995) - Pleckstrin Homology Domains: A Fact File. Current Opinion in Structural Biology, vol. 5, n. 3, p. 403–408. https://doi.org/10.1016/0959-440x(95)80104-9 [ Links ]
Schaub, R.E. & Dillard, J.P. (2019) - The Pathogenic Neisseria Use a Streamlined Set of Peptidoglycan Degradation Proteins for Peptidoglycan Remodeling, Recycling, and Toxic Fragment Release. Frontiers in Microbiology, vol. 10, art. 73. https://doi.org/10.3389/fmicb.2019.00073 [ Links ]
Schornack, S.; van Damme, M.; Bozkurt, T.O.; Cano, L.M.; Smoker, M.; Thines, M.; Gaulin, E.; Kamoun, S. & Huitema, E. (2010) - Ancient Class of Translocated Oomycete Effectors Targets the Host Nucleus. Proceedings of the National Academy of Sciences of the United States of America, vol. 107, n. 40, p. 17421–17426. https://doi.org/10.1073/pnas.1008491107 [ Links ]
Shattock, R. (2001) - Diseases of Woody Ornamentals and Trees in Nurseries. Vol. 50. John Wiley & Sons, Ltd. [ Links ]
Sigrist, C.J.A.; Cerutti, L.; de Castro, E.; Langendijk-Genevaux, P.S.; Bulliard, V.; Bairoch, A. & Hulo, N. (2009) - PROSITE, a Protein Domain Database for Functional Characterization and Annotation. Nucleic Acids Research, vol. 38, p. D161-D166. https://dx.doi.org/10.1093%2Fnar%2Fgkp885 [ Links ]
Silveira, D.R.; Milan, C.; Viana da Rosa, J. & Dias Timm, C. (2016) - Fatores de Patogenicidade de Vibrio Spp. de Importância Em Doenças Transmitidas Por Alimentos. Arquivos do Instituto Biológico, vol. 83, art. e1252013. http://dx.doi.org/10.1590/1808-1657001252013 [ Links ]
Singh, Y. & Gupta, R. (2016) - Novel S-Enantioselective Lipase TALipB from Trichosporon asahii MSR54: Heterologous Expression, Characterization, Conformational Stability and Homology Modeling. Enzyme and Microbial Technology, vol. 83, p. 29–39. https://doi.org/10.1016/j.enzmictec.2015.11.003 [ Links ]
Stam, R.; Jupe, J.; Howden, A.J.M.; Morris, J.A.; Boevink, P.C.; Hedley, P.E. & Huitema, E. (2013) - Identification and Characterisation CRN Effectors in Phytophthora capsici Shows Modularity and Functional Diversity. PLoS ONE, vol. 8, n. 3, art. e59517. https://doi.org/10.1371/journal.pone.0059517 [ Links ]
Studholme, D.J.; McDougal, R.L.; Sambles, C.; Hansen, E.; Hardy, G.; Grant, M.; Ganley, R.J. & Williams, N.M. (2016) - Genome Sequences of Six Phytophthora Species Associated with Forests in New Zealand. Genomics Data, vol. 7, p. 54–56. https://doi.org/10.1016/j.gdata.2015.11.015 [ Links ]
Thakare, H.S.; Meshram, D.B.; Jangam, C.M.; Labhasetwar, P.; Roychoudhary, K. & Ingle, A.B. (2016) - Comparative Genomics for Understanding the Structure, Function and Sub-Cellular Localization of Hypothetical Proteins in Thermanerovibrio acidaminovorans DSM 6589 (Tai). Computational Biology and Chemistry, vol. 61, p. 226–228. https://doi.org/10.1016/j.compbiolchem.2016.02.018 [ Links ]
Thanos, C.D.; Goodwill, K.E. & Bowie, J. U. (1999) - Oligomeric Structure of the Human EphB2 Receptor SAM Domain. Science, vol. 283, n. 5403, p. 833–836. https://doi.org/10.1126/science.283.5403.833 [ Links ]
Torto, T.A.; Li, S.; Styer, A.; Huitema, E.; Testa, A.; Gow, N.A.R.; van West, P. & Kamoun, S. (2003) - EST Mining and Functional Expression Assays Identify Extracellular Effector Proteins From the Plant Pathogen Phytophthora. Genome Research, vol. 13, n. 7, p. 1675–1685. https://doi.org/10.1101/gr.910003 [ Links ]
Vettraino, A.M.; Barzanti, G.P.; Bianco, M.C.; Ragazzi, A.; Capretti, P.; Paoletti, E.; Luisi, N.; Anselmi, N. & Vanninim A. (2002) - Occurrence of Phytophthora Species in Oak Stands in Italy and Their Association with Declining Oak Trees. Forest Pathology, vol. 32, n. 1, p. 19–28. https://doi.org/10.1046/j.1439-0329.2002.00264.x [ Links ]
Vettraino, A.M.; Natili, G.; Anselmi, N. & Vannini, A. (2001) - Recovery and Pathogenicity of Phytophthora Species Associated with a Resurgence of Ink Disease in Castanea sativa in Italy. Plant Pathology, vol. 50, n. 1, p. 90–96. https://doi.org/10.1046/j.1365-3059.2001.00528.x [ Links ]
Yokota, A.; Nishimura, K.; Takase, H.; Aoki, T.; Ogawa, T.; Tomizawa, K.I. & Ashida, H. (2009) - A Phosphofructokinase B-Type Carbohydrate Kinase Family Protein, NARA5, for Massive Expressions of Plastid-Encoded Photosynthetic Genes in Arabidopsis. Plant Physiology, vol. 151n. 1, p. 114–128. https://doi.org/10.1104/pp.109.139683 [ Links ]
Yu, C.-S.; Lin, C.-J. & Hwang, J.-K. (2004) - Predicting Subcellular Localization of Proteins for Gram-Negative Bacteria by Support Vector Machines Based on n -Peptide Compositions. Protein Science, vol. 13, n. 5, p. 1402–1406. https://doi.org/10.1110/ps.03479604 [ Links ]
Zhang, J.; Shang, Z.; Zhang, X. & Zhang, Y. (2011) - Modeling and Analysis of Schistosoma Argonaute Protein Molecular Spatial Conformation. Asian Pacific Journal of Tropical Biomedicine, vol. 1, n. 4, p. 275–278. https://dx.doi.org/10.1016%2FS2221-1691(11)60042-7 [ Links ]
ACKNOWLEDGMENT
The authors are grateful to Polytechnic Institute of Bragança (IPB) and especially to the Agricultural College of Bragança (IPB) for providing support to carry out this work.
Received/recebido: 2019.09.14
Accepted/aceite: 2019.11.29